Protein Structure Protein Structure 1 o The linear













- Slides: 73

Protein Structure

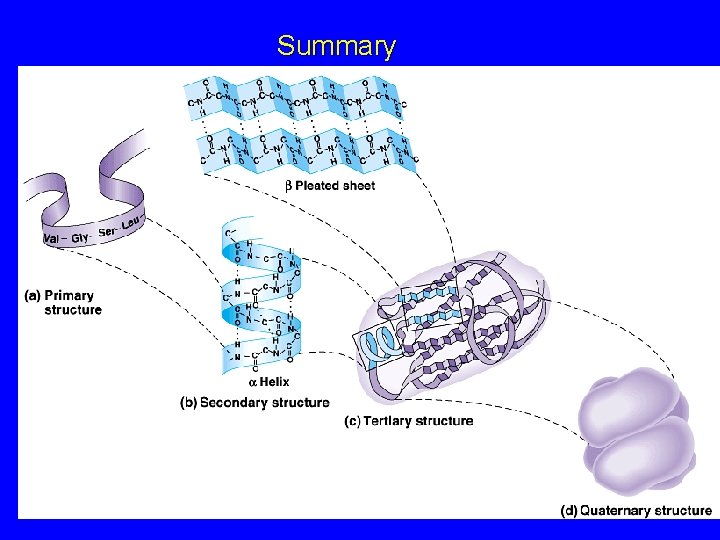
Protein Structure 1 o : The linear sequence of amino acids and disulfide bonds eg. ARDV: Ala. Arg. Asp. Val. 2 o : Local structures which include, folds, turns, -helices and -sheets held in place by hydrogen bonds. 3 o : 3 -D arrangement of all atoms in a single polypeptide chain. 4 o : Arrangement of polypeptide chains into a functional protein, eg. hemoglobin.

Protein Structure 1 o : The linear sequence of amino acids and disulfide bonds eg. ARDV: Ala. Arg. Asp. Val. 2 o : Local structures which include, folds, turns, -helices and -sheets held in place by hydrogen bonds. 3 o : 3 -D arrangement of all atoms in a single polypeptide chain. 4 o : Arrangement of polypeptide chains into a functional protein, eg. hemoglobin.

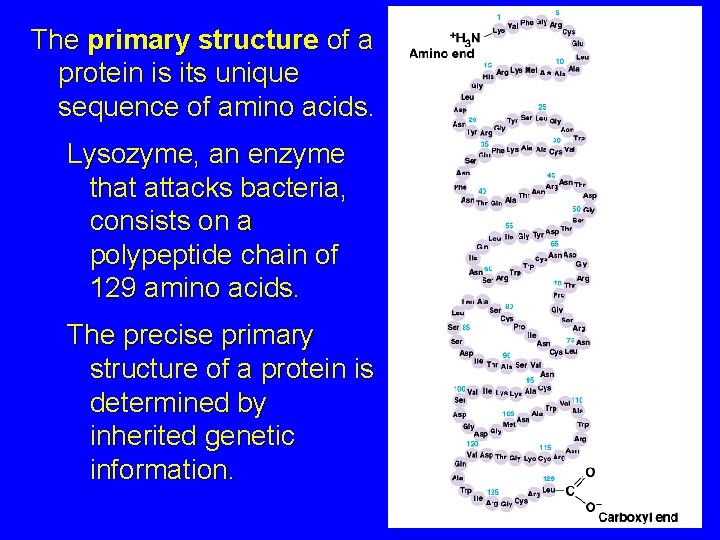
The primary structure of a protein is its unique sequence of amino acids. Lysozyme, an enzyme that attacks bacteria, consists on a polypeptide chain of 129 amino acids. The precise primary structure of a protein is determined by inherited genetic information.

Protein Structure 1 o : The linear sequence of amino acids and disulfide bonds eg. ARDV: Ala. Arg. Asp. Val. 2 o : Local structures which include, folds, turns, -helices and -sheets held in place by hydrogen bonds. 3 o : 3 -D arrangement of all atoms in a single polypeptide chain. 4 o : Arrangement of polypeptide chains into a functional protein, eg. hemoglobin.

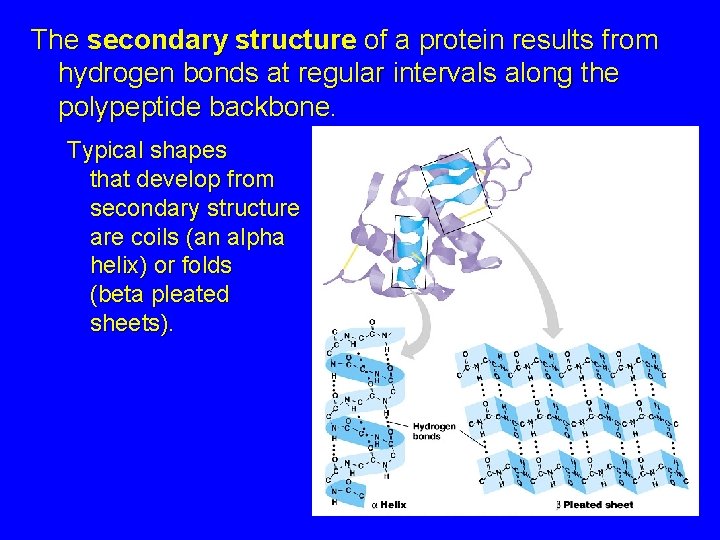
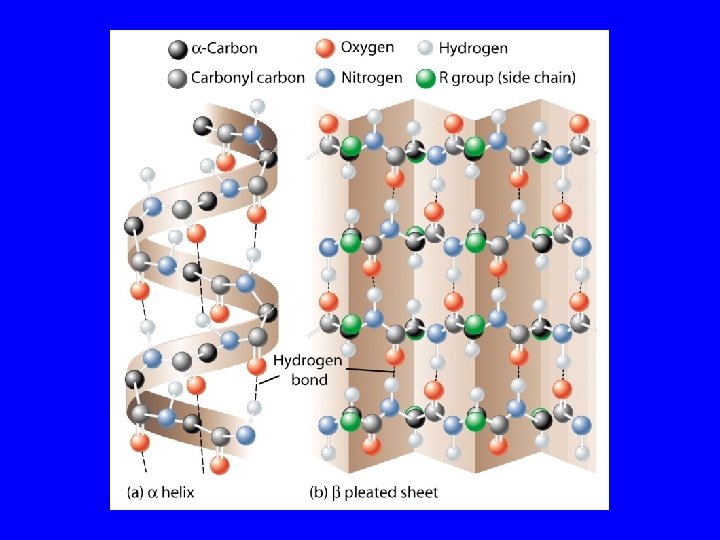
The secondary structure of a protein results from hydrogen bonds at regular intervals along the polypeptide backbone. Typical shapes that develop from secondary structure are coils (an alpha helix) or folds (beta pleated sheets).

The structural properties of silk are due to beta pleated sheets. The presence of so many hydrogen bonds makes each silk fiber stronger than steel.

Fibrous Proteins Fibrous proteins have high a-helix or -sheet content. Most are structural proteins. Examples include: collagen elastin keratin


Collagen - A Triple Helix Principal component of connective tissue (tendons, cartilage, bones, teeth) Basic unit is tropocollagen: three intertwined polypeptide chains (1000 residues each MW = 285, 000 300 nm long, 1. 4 nm diameter unique amino acid composition

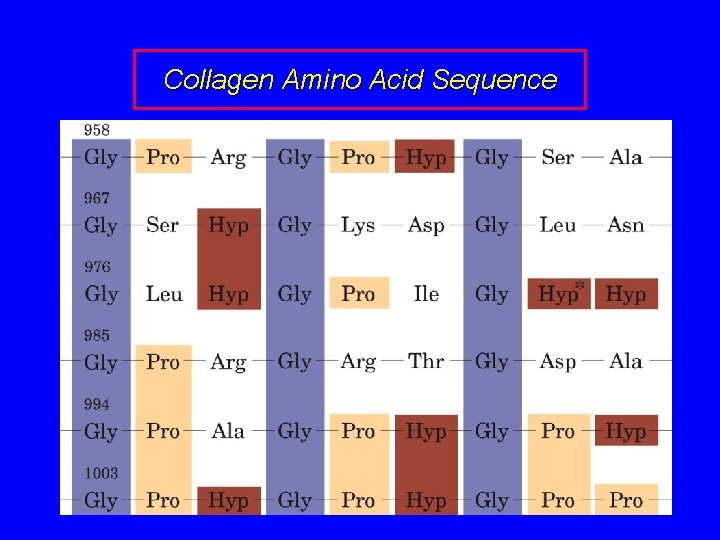
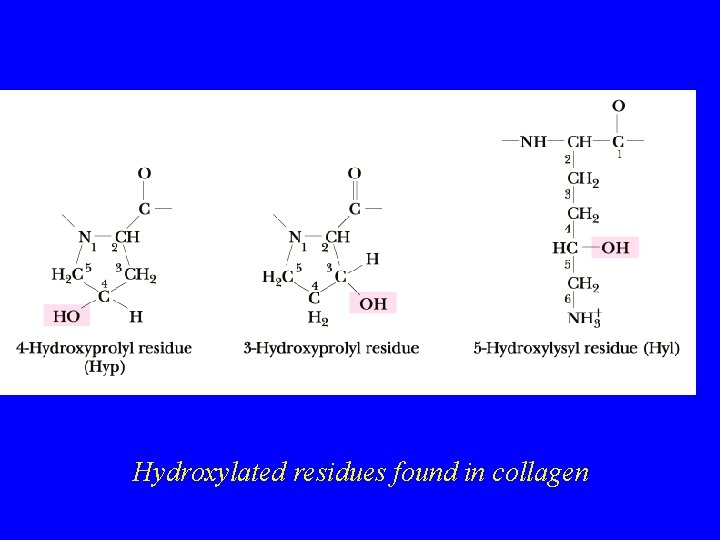
Collagen Amino Acid Composition Nearly one residue out of three is Gly Proline content is unusually high Many modified amino acids present: 4 -hydroxyproline 3 -hydroxyproline 5 -hydroxylysine Pro and Hy. Pro together make 30% of res.

Collagen Amino Acid Sequence

Hydroxylated residues found in collagen

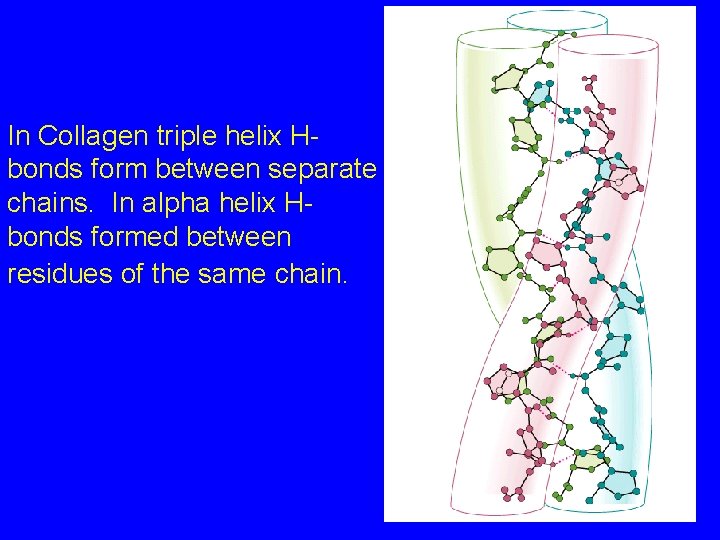
The Collagen Triple Helix The unusual amino acid composition of collagen is not favorable for alpha helices OR beta sheets But it is ideally suited for the collagen triple helix: three intertwined helical strands Much more extended than alpha helix, with a rise per residue of 2. 9 Angstroms 3. 3 residues per turn Long stretches of Gly-Pro/Hy. P

In Collagen triple helix Hbonds form between separate chains. In alpha helix Hbonds formed between residues of the same chain.

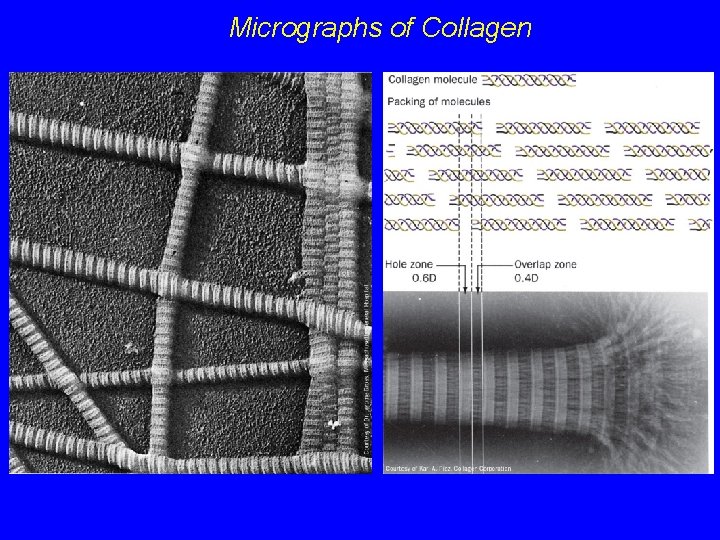
Collagen Fibers are formed by staggered arrays of tropocollagens Banding pattern in EMs with 68 nm repeat Since tropocollagens are 300 nm long, there must be 40 nm gaps between adjacent tropocollagens (5 x 68 = 340 Angstroms) 40 nm gaps are called "hole regions" - they contain carbohydrate and are thought to be nucleation sites for bone formation

Electron Micrographs of Collagen fibers showing b

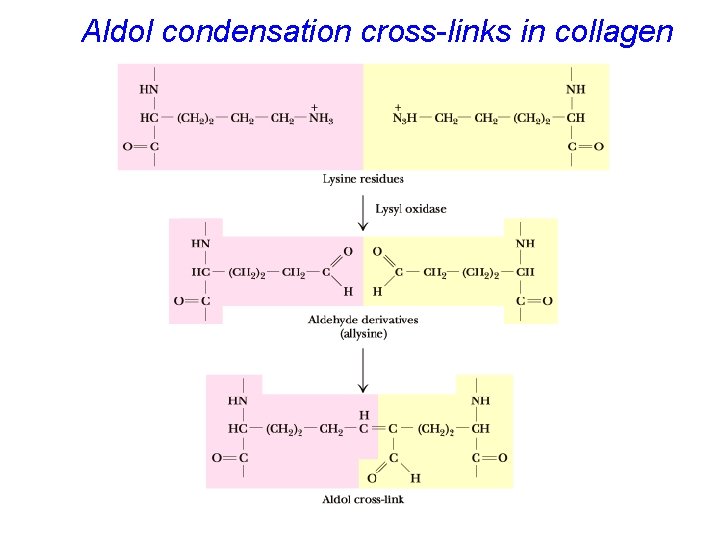
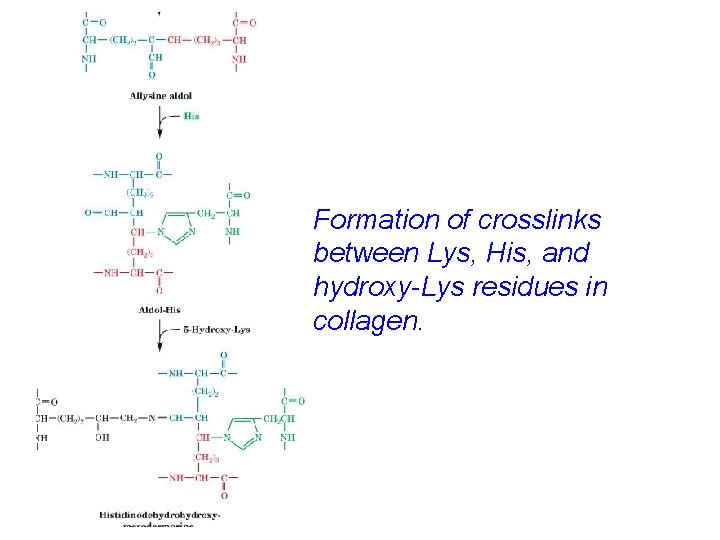
Structural basis of the collagen triple helix Every third residue faces the crowded center of the helix - only Gly fits Pro and Hy. P suit the constraints of phi and psi Interchain H-bonds involving Hy. P stabilize helix Fibrils are strengthened by intrachain lysine-lysine and interchain hydroxypyridinium crosslinks

Aldol condensation cross-links in collagen

Formation of crosslinks between Lys, His, and hydroxy-Lys residues in collagen.

Elastin Abundant in ligaments, lungs, artery walls, skin. Provides tissues with ability to stretch in all directions without tearing. Contains predominantly small hydrophobic residues: 1/3 Gly, 1/3 Ala + Val, many Pro but no hydroxy. Pro or hydroxy. Lys. Lacks regular secondary structure. Has unordered coil structure that is highly cross-linked into 3 -dimensional network of fibers to provide rubber -like elasticity.

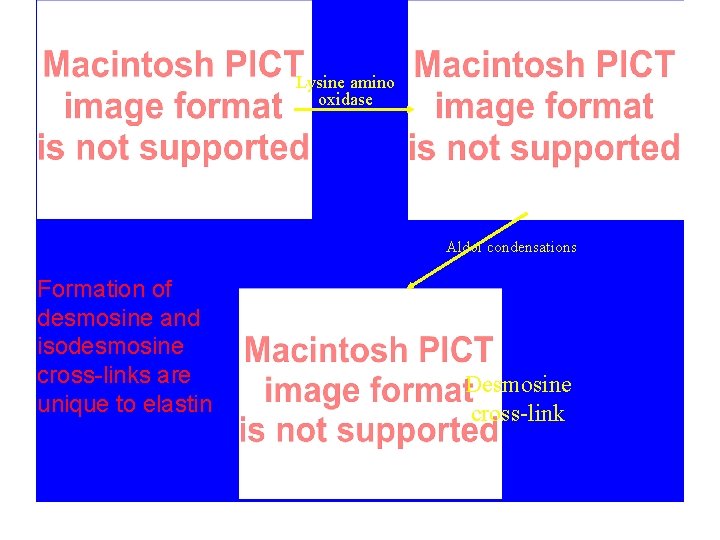
Elastin Cross-links formed from allyysine (aldehyde derivative of Lys) Extracellular lysine amino oxidase specific for Lys-Ala. Ala-Lys and Lys-(Ala)3 -Lys sequences Lys + 3 allysine combine to from desmosine or isodesmosine cross-links responsible for yellow color of elastin Also forms lysine or leucine cross-links from 2 allysine, as in collagen. Cross-links responsible for elasticity & insolubility

Lysine amino oxidase Aldol condensations Formation of desmosine and isodesmosine cross-links are unique to elastin Desmosine cross-link

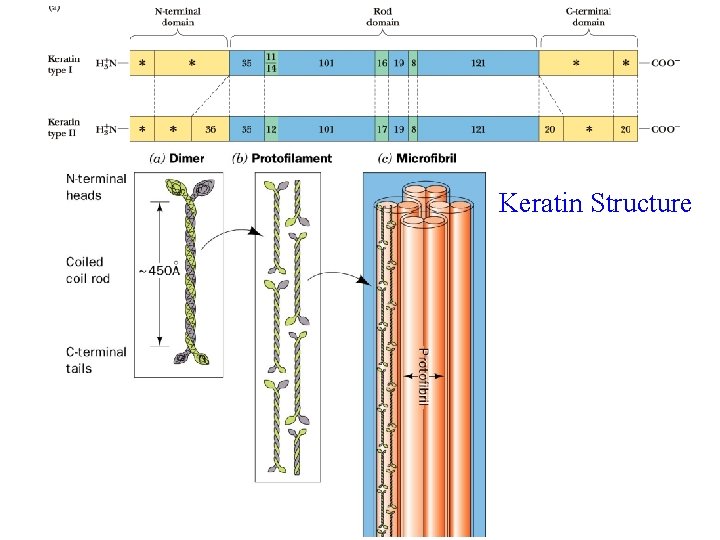
Alpha Keratin Found in hair, fingernails, claws, horns and beaks Sequence consists of 311 -314 residue alpha helical segments capped with non-helical N- and C-termini Primary structure of helical rods consists of 7 -residue repeats: (a-b-c-d-e-f-g)n, where a and d are nonpolar. Promotes association of helices!

Keratin Structure


Protein Structure 1 o : The linear sequence of amino acids and disulfide bonds eg. ARDV: Ala. Arg. Asp. Val. 2 o : Local structures which include, folds, turns, -helices and -sheets held in place by hydrogen bonds. 3 o : 3 -D arrangement of all atoms in a single polypeptide chain. 4 o : Arrangement of polypeptide chains into a functional protein, eg. hemoglobin.

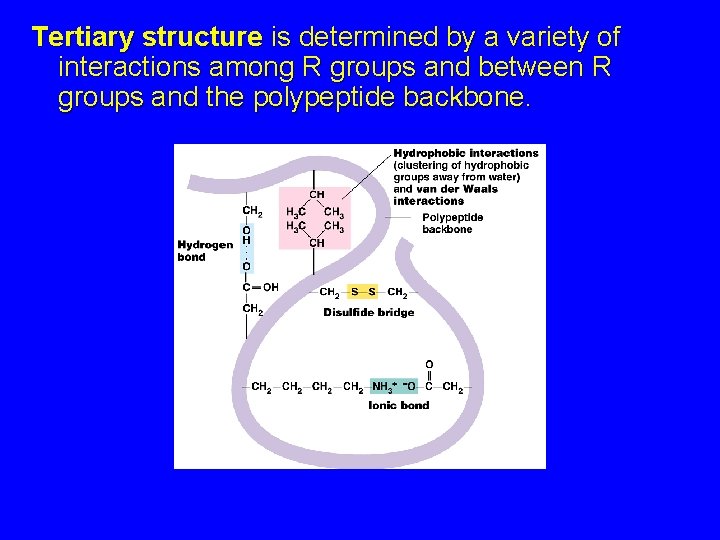
Tertiary structure is determined by a variety of interactions among R groups and between R groups and the polypeptide backbone.

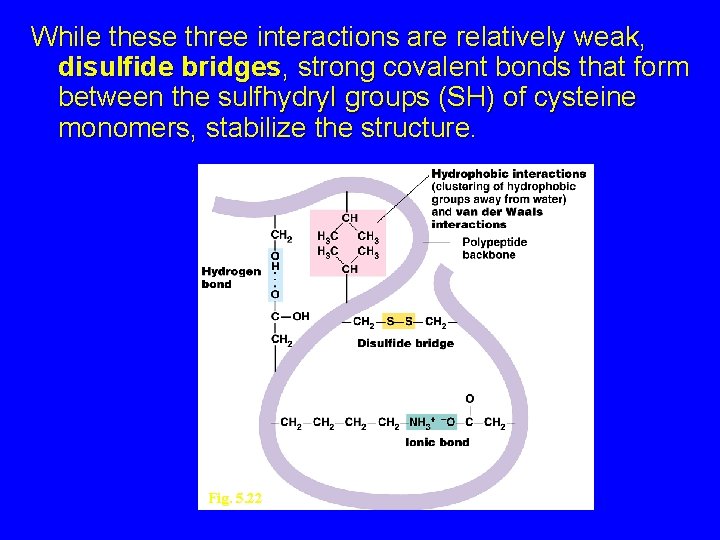
While these three interactions are relatively weak, disulfide bridges, strong covalent bonds that form between the sulfhydryl groups (SH) of cysteine monomers, stabilize the structure. Fig. 5. 22

Protein Structure 1 o : The linear sequence of amino acids and disulfide bonds eg. ARDV: Ala. Arg. Asp. Val. 2 o : Local structures which include, folds, turns, -helices and -sheets held in place by hydrogen bonds. 3 o : 3 -D arrangement of all atoms in a single polypeptide chain. 4 o : Arrangement of polypeptide chains into a functional protein, eg. hemoglobin.

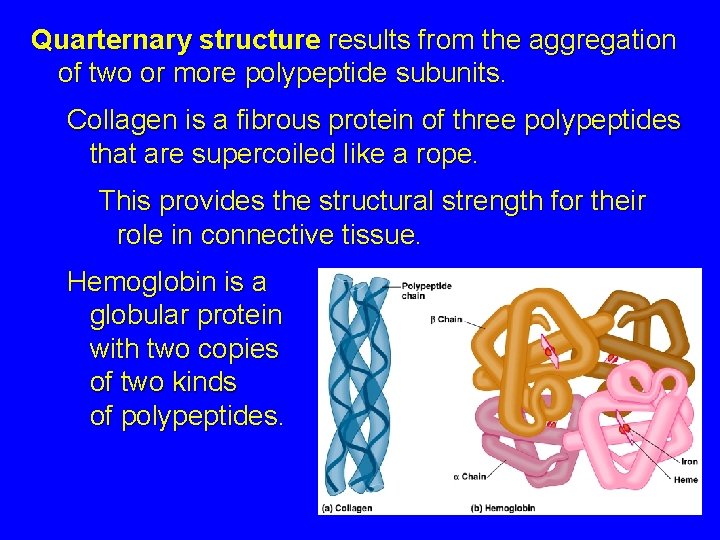
Quarternary structure results from the aggregation of two or more polypeptide subunits. Collagen is a fibrous protein of three polypeptides that are supercoiled like a rope. This provides the structural strength for their role in connective tissue. Hemoglobin is a globular protein with two copies of two kinds of polypeptides.


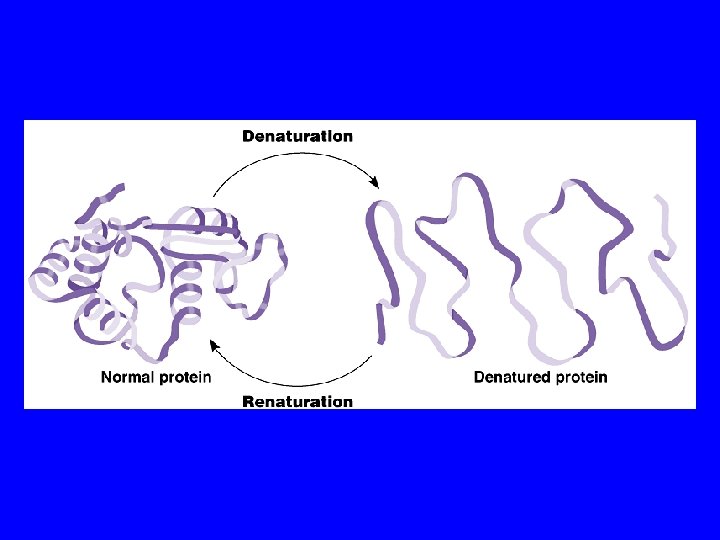
A protein’s conformation can change in response to physical and/or chemical conditions. Alterations in p. H, salt concentration, temperature, or other factors such as the presence of O 2 can alter (an allosteric effect), or unravel, or denature a protein. These forces alter the hydrogen bonds, ionic bonds, and disulfide bridges that maintain the protein’s shape. Some proteins can return to their functional shape such as hemoglobin, but others cannot, eg. the white of a cooked egg.



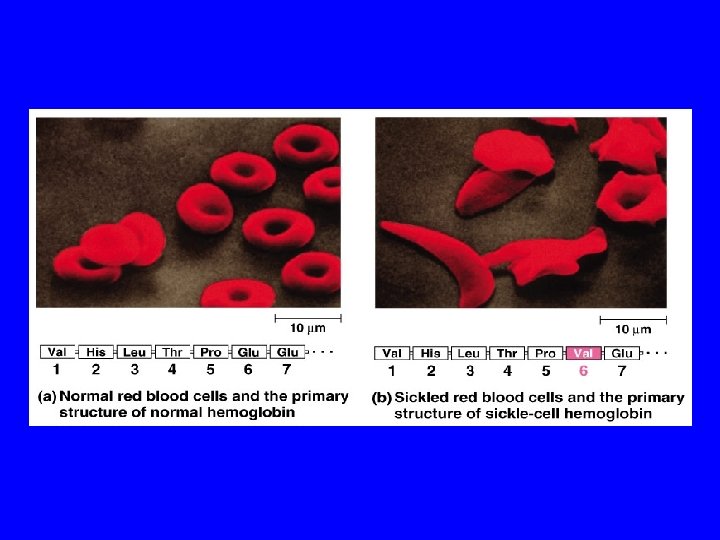
Even a slight change in primary structure can affect a protein’s conformation and ability to function. In individuals with sickle cell disease, abnormal hemoglobins, oxygen-carrying proteins, develop because of a single amino acid substitution. These abnormal hemoglobins crystallize, deforming the red blood cells and leading to clogs in tiny blood vessels.

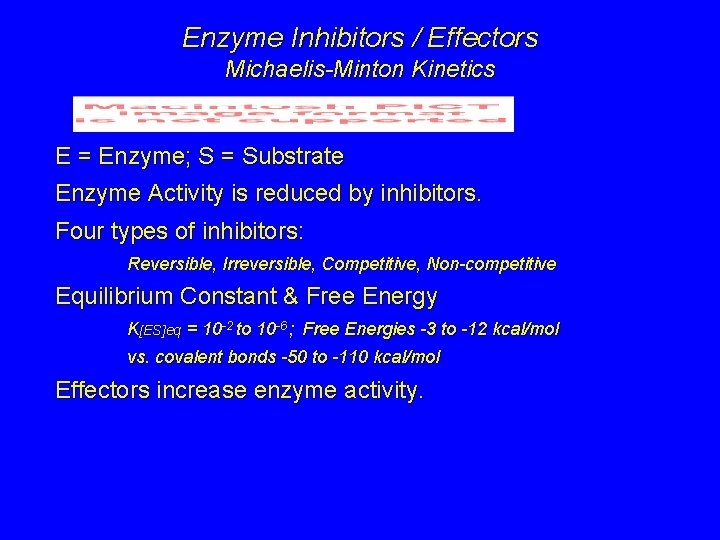
Enzyme Inhibitors / Effectors Michaelis-Minton Kinetics E = Enzyme; S = Substrate Enzyme Activity is reduced by inhibitors. Four types of inhibitors: Reversible, Irreversible, Competitive, Non-competitive Equilibrium Constant & Free Energy K[ES]eq = 10 -2 to 10 -6 ; Free Energies -3 to -12 kcal/mol vs. covalent bonds -50 to -110 kcal/mol Effectors increase enzyme activity.

Acetylcholinesterase Docking

Coenzymes The range of chemical reactions that amino acid side chains can participate in is relatively limited. acid-base (transfer and accepting protons)substitution Many other biological processes, such as oxidationreduction, require coenzymes, cofactors, or prosthetic groups in order to occur.

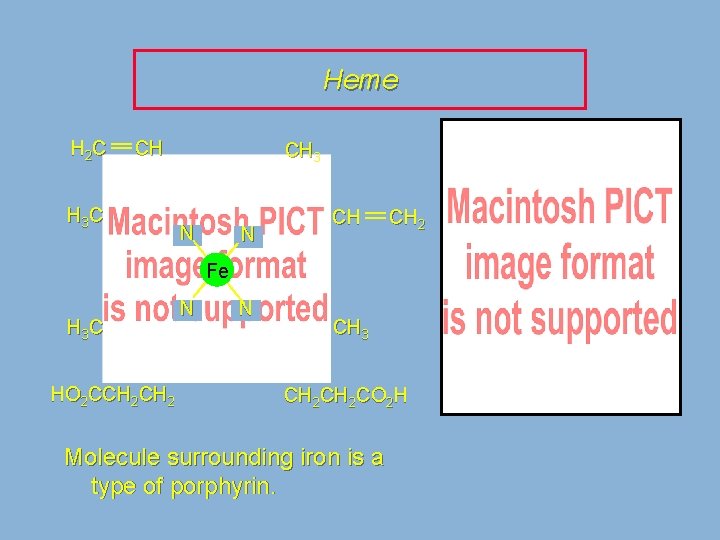
Coenzymes NADH, coenzyme A and coenzyme B 12 are examples of coenzymes. Heme is another example.

Heme H 2 C CH H 3 C CH 3 N N CH CH 2 Fe H 3 C HO 2 CCH 2 N N CH 3 CH 2 CO 2 H Molecule surrounding iron is a type of porphyrin.

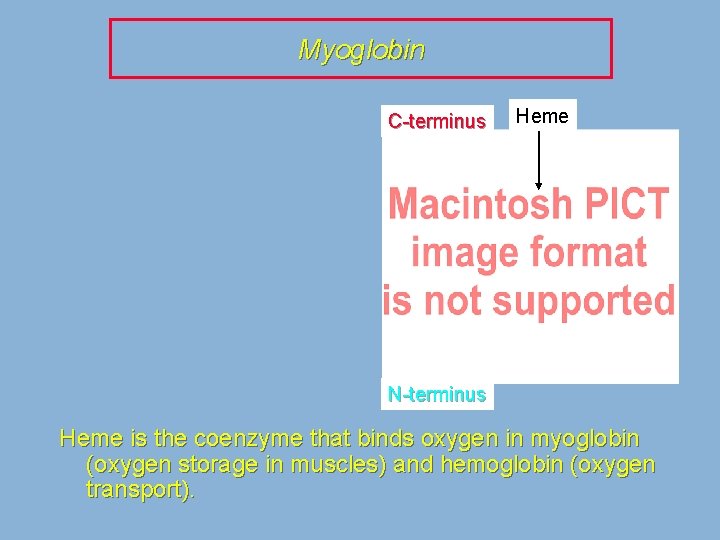
Myoglobin C-terminus Heme N-terminus Heme is the coenzyme that binds oxygen in myoglobin (oxygen storage in muscles) and hemoglobin (oxygen transport).

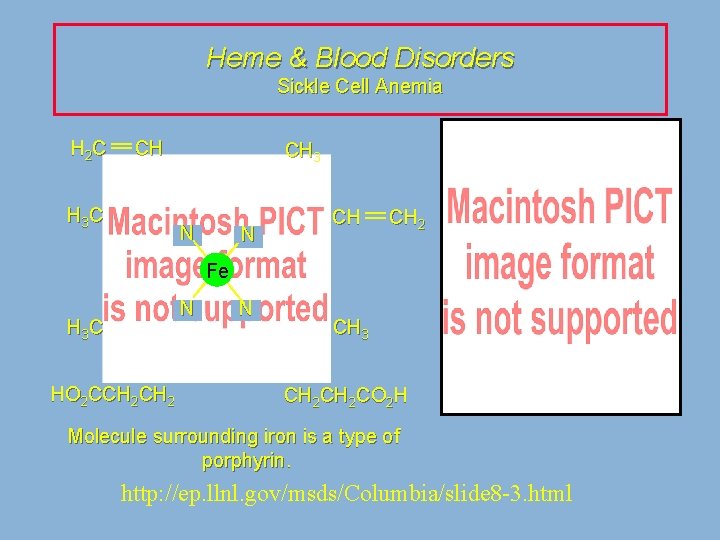
Heme & Blood Disorders Sickle Cell Anemia H 2 C CH H 3 C CH 3 N N CH CH 2 Fe N H 3 C HO 2 CCH 2 N CH 3 CH 2 CO 2 H Molecule surrounding iron is a type of porphyrin. http: //ep. llnl. gov/msds/Columbia/slide 8 -3. html

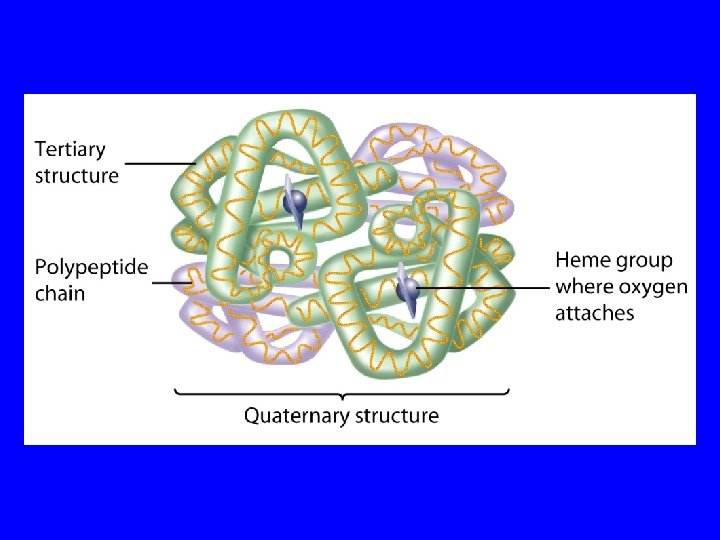
Protein Quaternary Structure Some proteins are assemblies of two or more chains. The way in which these chains are organized is called the quaternary structure. Hemoglobin, for example, consists of 4 subunits. There are 2 a chains (identical) and 2 chains (also identical). Each subunit contains one heme and each protein is about the size of myoglobin.

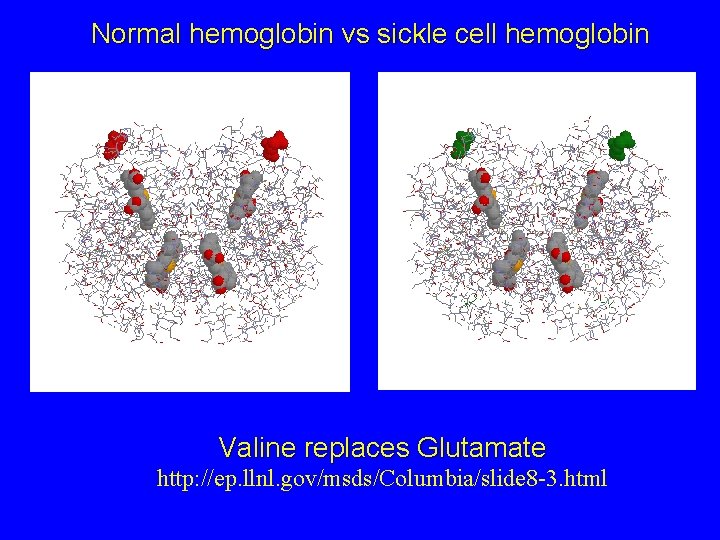
Normal hemoglobin vs sickle cell hemoglobin Valine replaces Glutamate http: //ep. llnl. gov/msds/Columbia/slide 8 -3. html

Summary

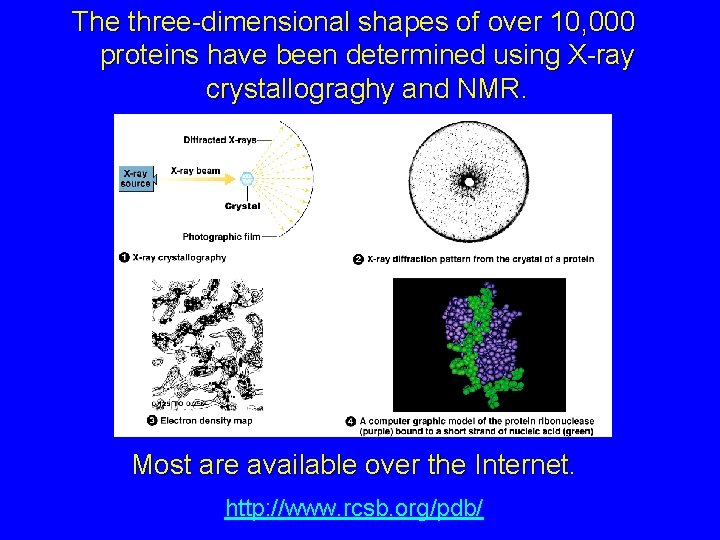
The three-dimensional shapes of over 10, 000 proteins have been determined using X-ray crystallograghy and NMR. Most are available over the Internet. http: //www. rcsb. org/pdb/

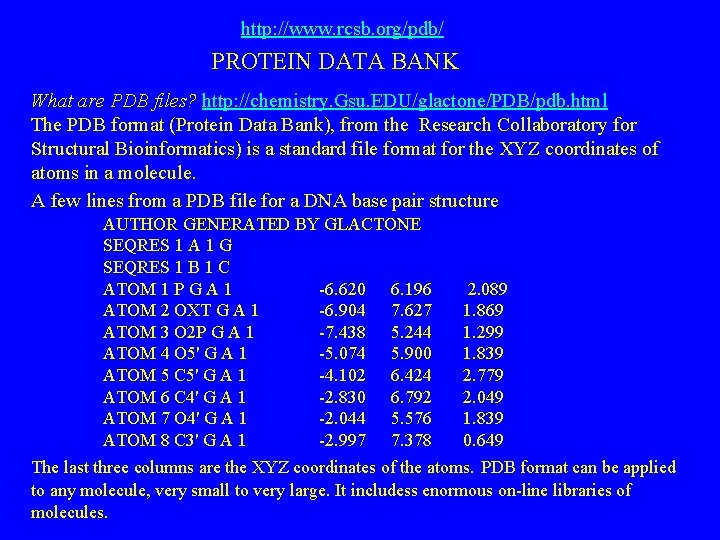
http: //www. rcsb. org/pdb/ PROTEIN DATA BANK What are PDB files? http: //chemistry. Gsu. EDU/glactone/PDB/pdb. html The PDB format (Protein Data Bank), from the Research Collaboratory for Structural Bioinformatics) is a standard file format for the XYZ coordinates of atoms in a molecule. A few lines from a PDB file for a DNA base pair structure AUTHOR GENERATED BY GLACTONE SEQRES 1 A 1 G SEQRES 1 B 1 C ATOM 1 P G A 1 -6. 620 6. 196 ATOM 2 OXT G A 1 -6. 904 7. 627 ATOM 3 O 2 P G A 1 -7. 438 5. 244 ATOM 4 O 5' G A 1 -5. 074 5. 900 ATOM 5 C 5' G A 1 -4. 102 6. 424 ATOM 6 C 4' G A 1 -2. 830 6. 792 ATOM 7 O 4' G A 1 -2. 044 5. 576 ATOM 8 C 3' G A 1 -2. 997 7. 378 2. 089 1. 869 1. 299 1. 839 2. 779 2. 049 1. 839 0. 649 The last three columns are the XYZ coordinates of the atoms. PDB format can be applied to any molecule, very small to very large. It includess enormous on-line libraries of molecules.

Proteins Self Assemble http: //www. stark. kent. edu/~cearley/PChem/protein. htm

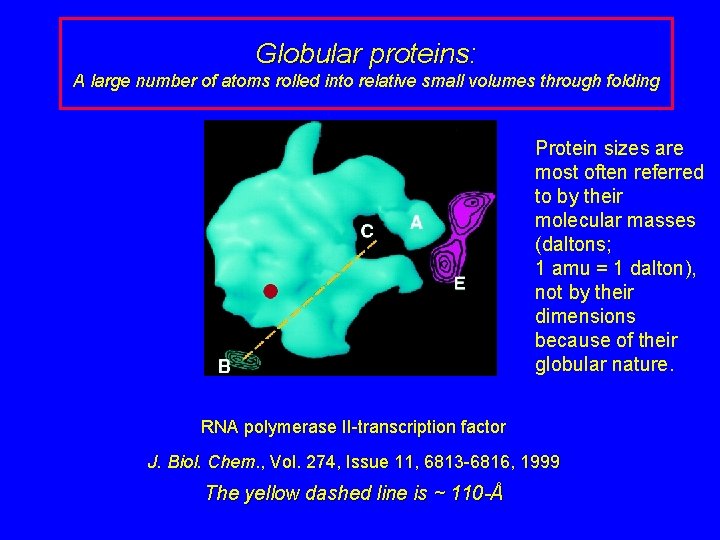
Globular proteins: A large number of atoms rolled into relative small volumes through folding Protein sizes are most often referred to by their molecular masses (daltons; 1 amu = 1 dalton), not by their dimensions because of their globular nature. RNA polymerase II-transcription factor J. Biol. Chem. , Vol. 274, Issue 11, 6813 -6816, 1999 The yellow dashed line is ~ 110 -Å

Although the three-dimensional shapes of over 10, 000 proteins are known, it is difficult to predict the conformation of a protein from its primary structure. Proteins fold and undergo changes. There are dynamic forms in addition to a “mature” conformation found in crystals. Supercomputers and distributed computing are being used to predict folding patterns and generate the conformation of any protein from its primary structure (amino acid sequence) or its DNA/gene sequence. Want to participate? http: //folding. stanford. edu/

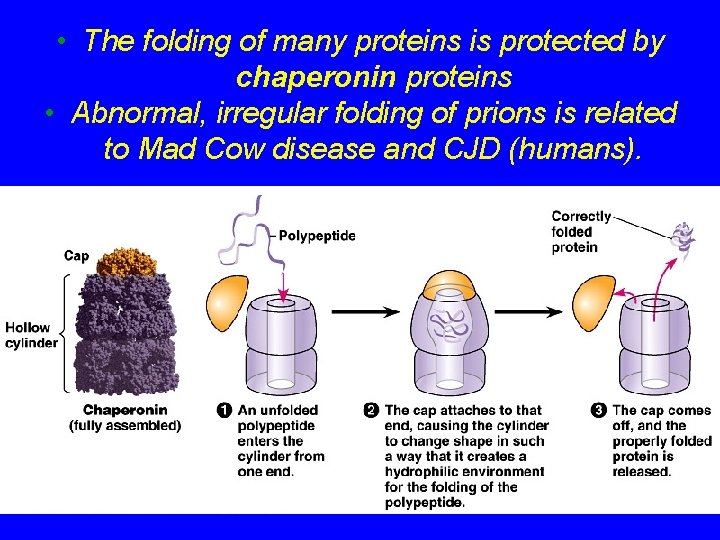
• The folding of many proteins is protected by chaperonin proteins • Abnormal, irregular folding of prions is related to Mad Cow disease and CJD (humans).

The Importance of Accurate Prediction Enzyme Interactions: eg. Neurotransmission The interaction of a globular protein, acetylcholinesterase, with a relatively small molecule, acetylcholine. Richard Short (Cornell University)

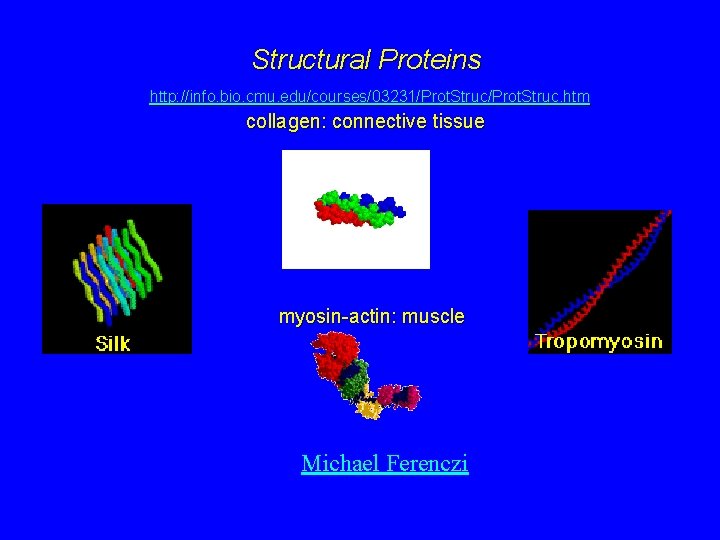
Structural Proteins http: //info. bio. cmu. edu/courses/03231/Prot. Struc. htm collagen: connective tissue myosin-actin: muscle Michael Ferenczi

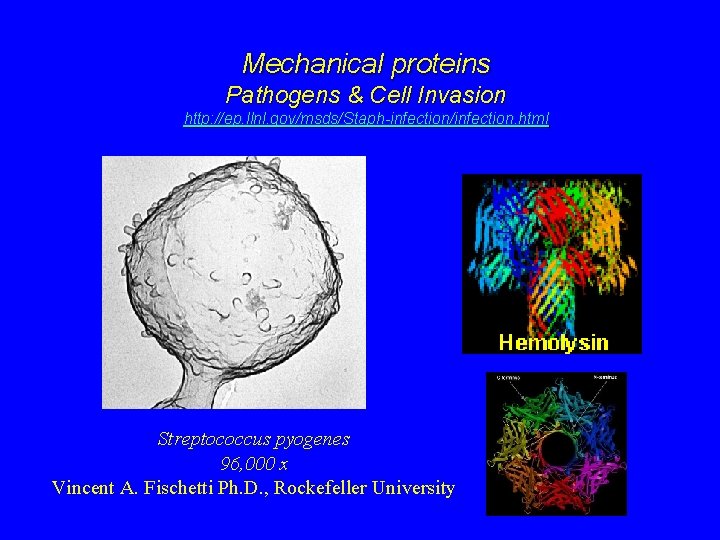
Mechanical proteins Pathogens & Cell Invasion http: //ep. llnl. gov/msds/Staph-infection/infection. html Streptococcus pyogenes 96, 000 x Vincent A. Fischetti Ph. D. , Rockefeller University

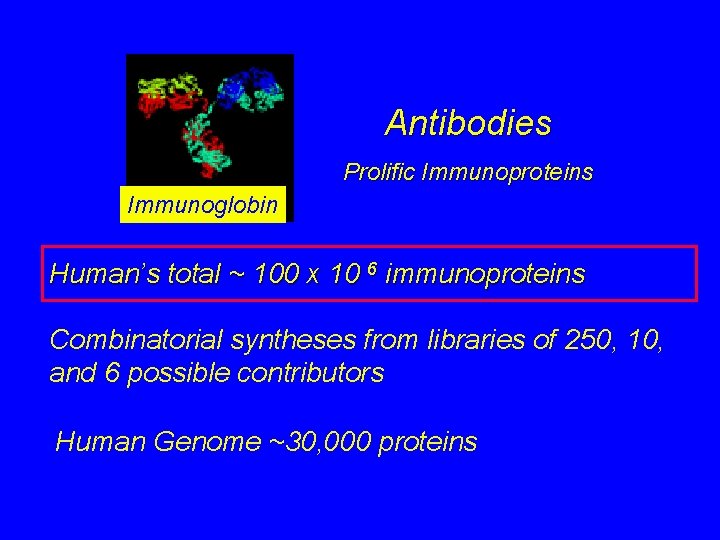
Antibodies Prolific Immunoproteins Immunoglobin Human’s total ~ 100 x 10 6 immunoproteins Combinatorial syntheses from libraries of 250, 10, and 6 possible contributors Human Genome ~30, 000 proteins

Some Processes Used in Protein Structure Determination Molecular exclusion/gel filtration chromatography Amino Acid Sequencing Mass Spectrometry

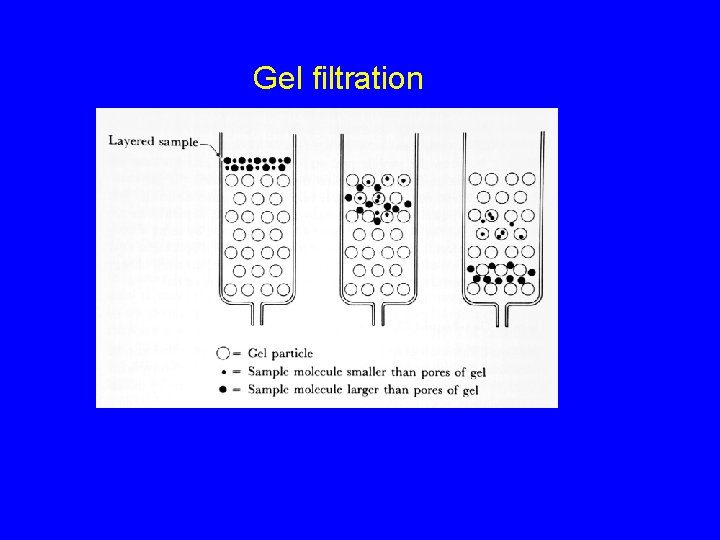
Gel filtration

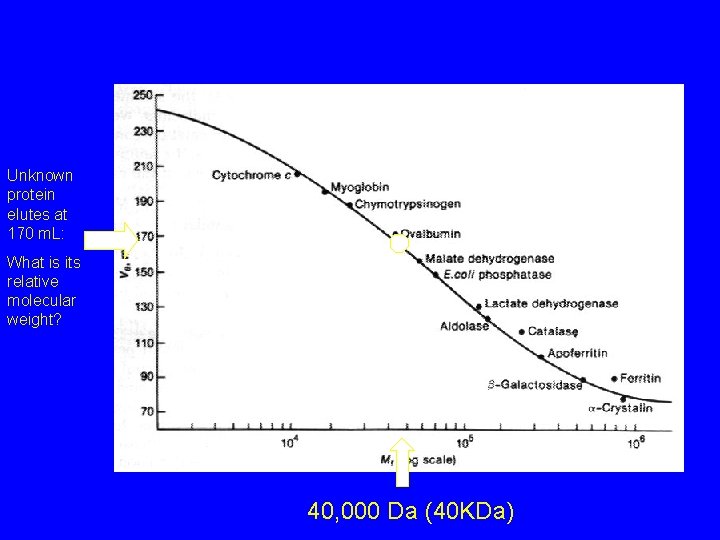
Unknown protein elutes at 170 m. L: What is its relative molecular weight? 40, 000 Da (40 KDa)

Some Processes Used in Protein Structure Determination Molecular exclusion/gel filtration chromatography Amino Acid Sequencing Mass Spectrometry

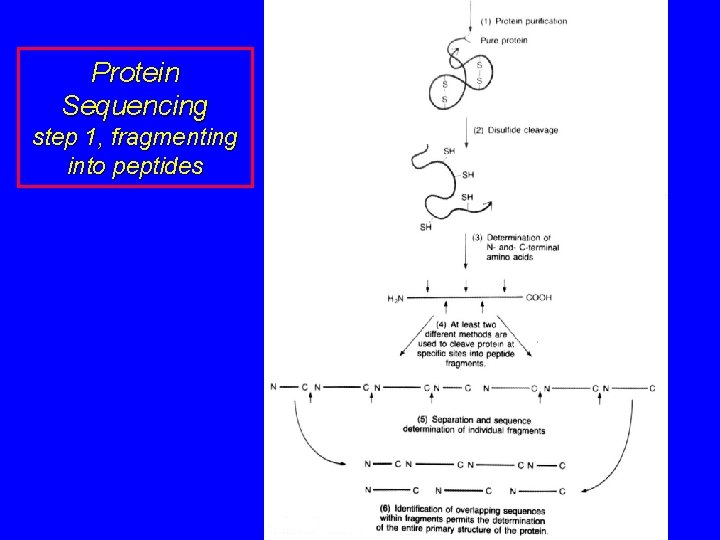
Protein Sequencing step 1, fragmenting into peptides

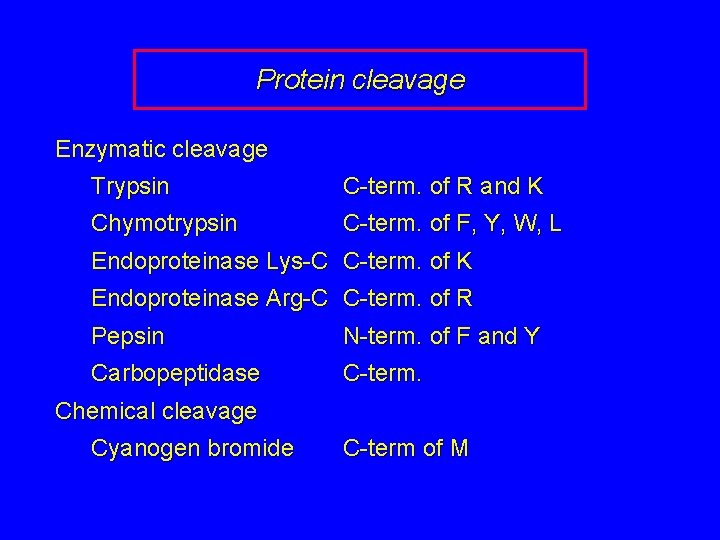
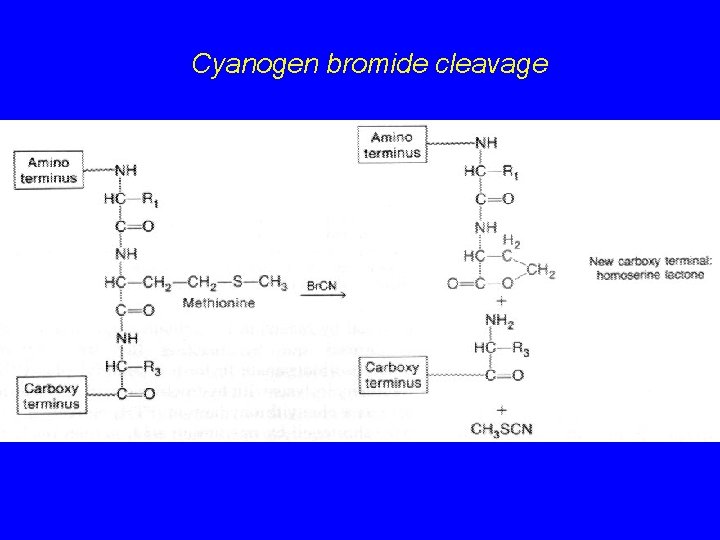
Protein cleavage Enzymatic cleavage Trypsin C-term. of R and K Chymotrypsin C-term. of F, Y, W, L Endoproteinase Lys-C C-term. of K Endoproteinase Arg-C C-term. of R Pepsin N-term. of F and Y Carbopeptidase C-term. Chemical cleavage Cyanogen bromide C-term of M

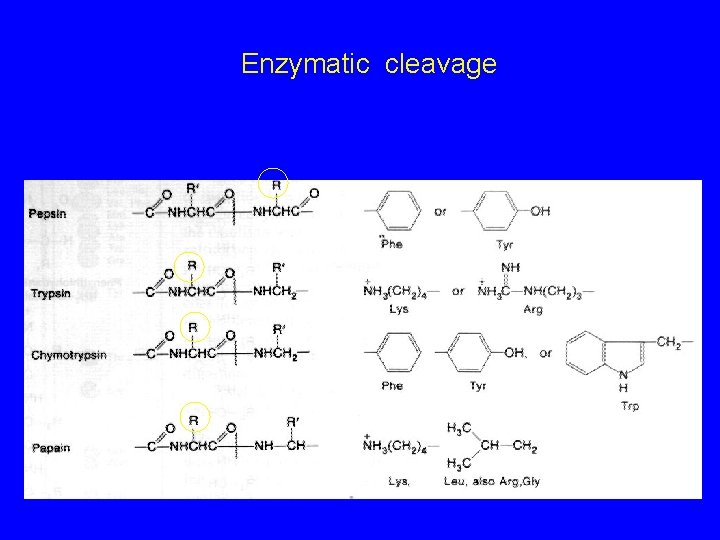
Enzymatic cleavage

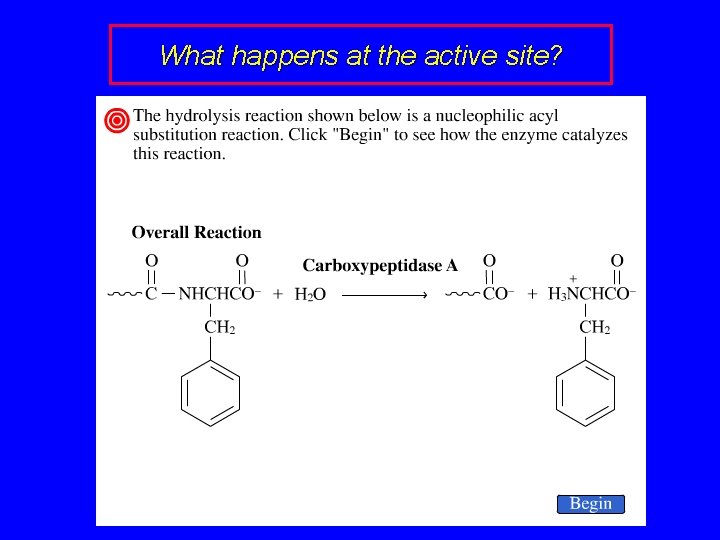
Carboxypeptidase is an enzyme that catalyzes the hydrolysis of proteins at their C-terminus. It is a metalloenzyme containing Zn 2+ at its active site. An amino acid with a positively charged side chain (Arg-145) is near the active site.

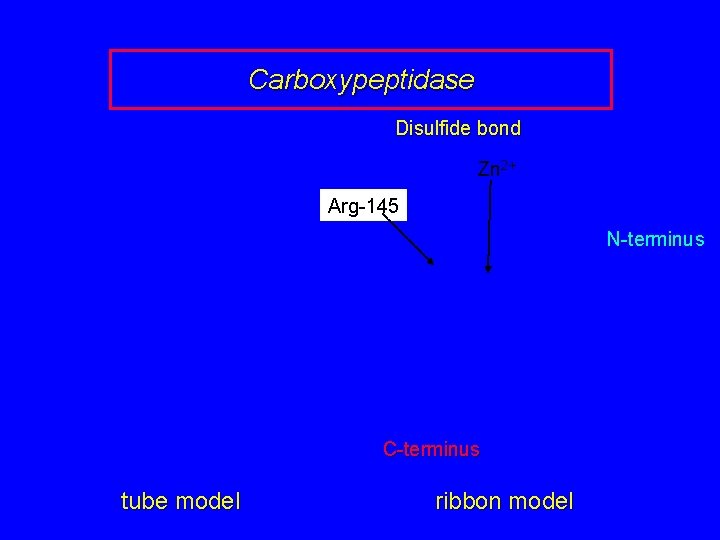
Carboxypeptidase Disulfide bond Zn 2+ Arg-145 N-terminus C-terminus tube model ribbon model

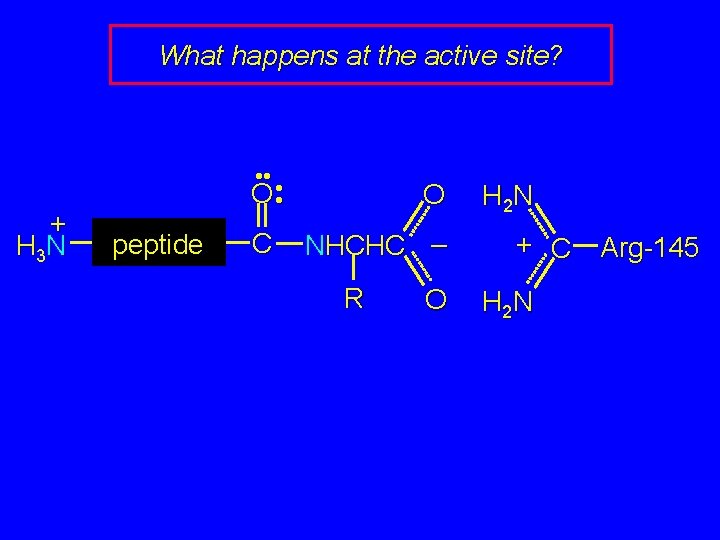
What happens at the active site?

What happens at the active site? + H 3 N • • • O • peptide C O NHCHC – R O H 2 N + C H 2 N Arg-145

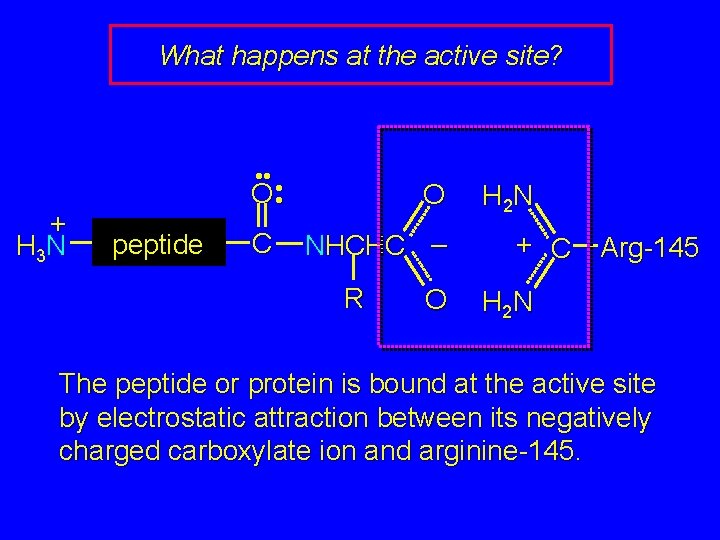
What happens at the active site? + H 3 N • • • O • peptide C O NHCHC – R O H 2 N + C Arg-145 H 2 N The peptide or protein is bound at the active site by electrostatic attraction between its negatively charged carboxylate ion and arginine-145.

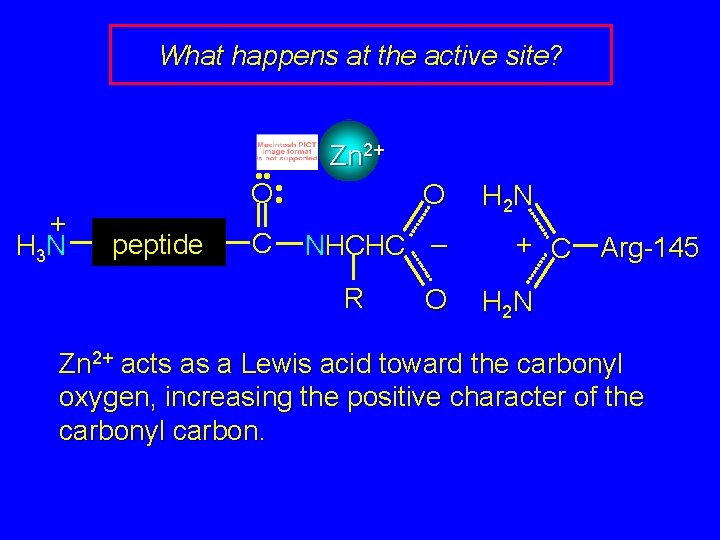
What happens at the active site? + H 3 N • • • O • peptide C Zn 2+ O NHCHC – R O H 2 N + C Arg-145 H 2 N Zn 2+ acts as a Lewis acid toward the carbonyl oxygen, increasing the positive character of the carbonyl carbon.

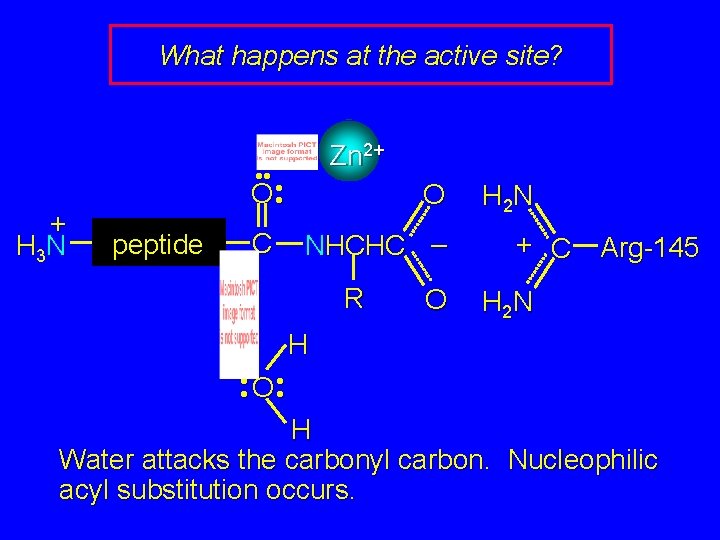
What happens at the active site? + H 3 N Zn 2+ • • • O • peptide C O NHCHC – R O H 2 N + C Arg-145 H 2 N H • O • • • H Water attacks the carbonyl carbon. Nucleophilic acyl substitution occurs.

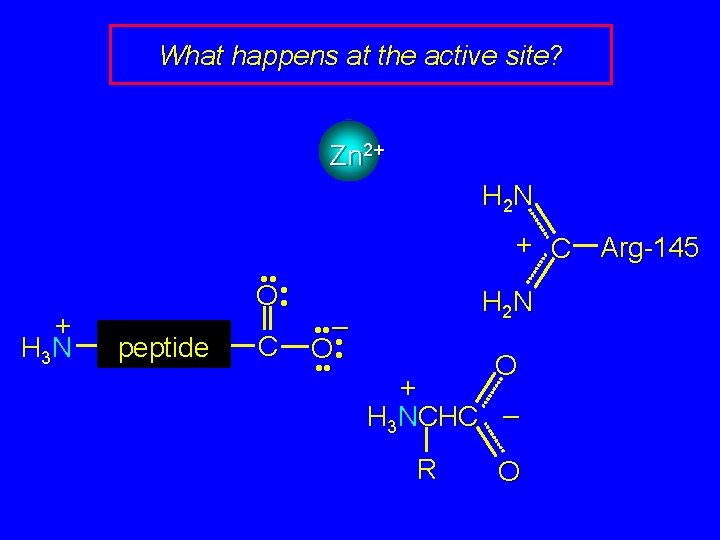
What happens at the active site? Zn 2+ H 2 N + C + H 3 N • • • O • peptide C • • – O • • H 2 N O + H 3 NCHC – R O Arg-145

Cyanogen bromide cleavage

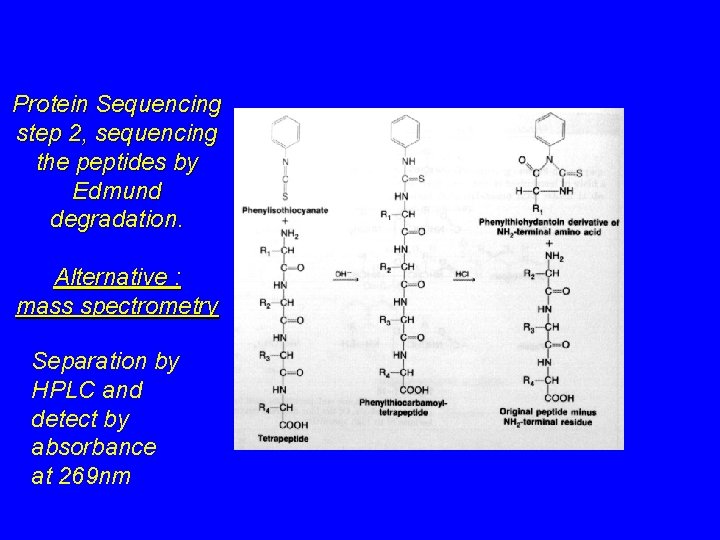
Protein Sequencing step 2, sequencing the peptides by Edmund degradation. Alternative : mass spectrometry Separation by HPLC and detect by absorbance at 269 nm