Protein Structure Prediction Protein Structure u Aminoacid chains










- Slides: 52

Protein Structure Prediction .

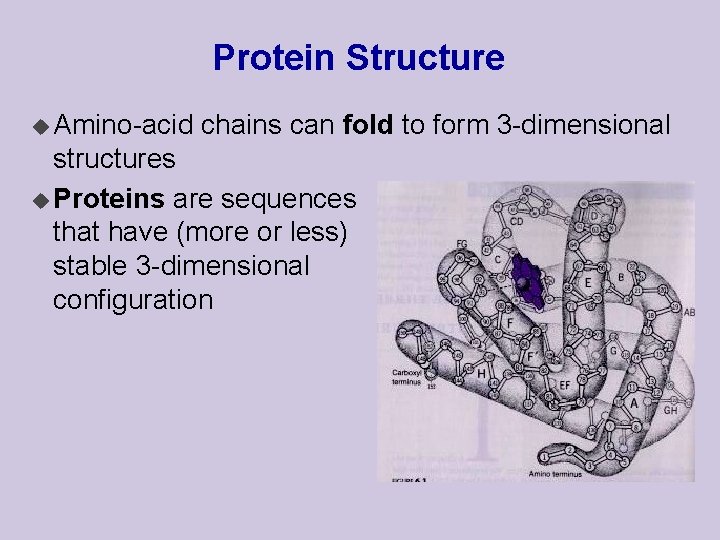
Protein Structure u Amino-acid chains can fold to form 3 -dimensional structures u Proteins are sequences that have (more or less) stable 3 -dimensional configuration

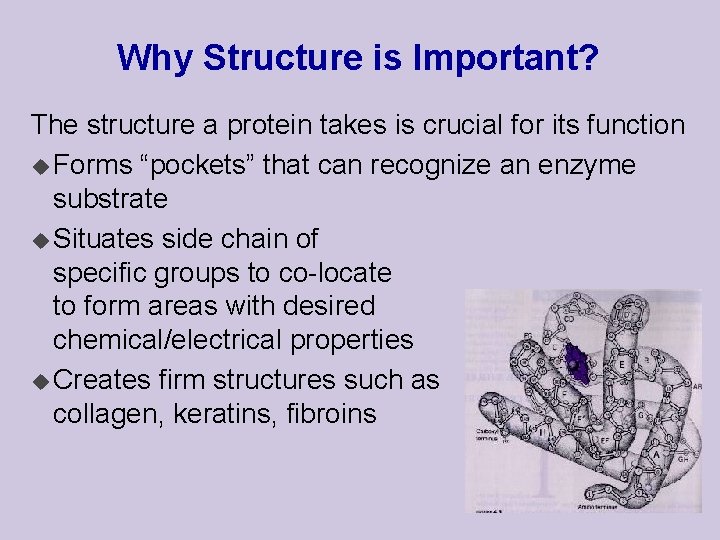
Why Structure is Important? The structure a protein takes is crucial for its function u Forms “pockets” that can recognize an enzyme substrate u Situates side chain of specific groups to co-locate to form areas with desired chemical/electrical properties u Creates firm structures such as collagen, keratins, fibroins

Determining Structure u X-Ray and NMR methods allow to determine the structure of proteins and protein complexes u These methods are expensive and difficult l Could take several work months to process one proteins u. A centralized database (PDB) contains all solved protein structures l XYZ coordinate of atoms within specified precision l ~19, 000 solved structures

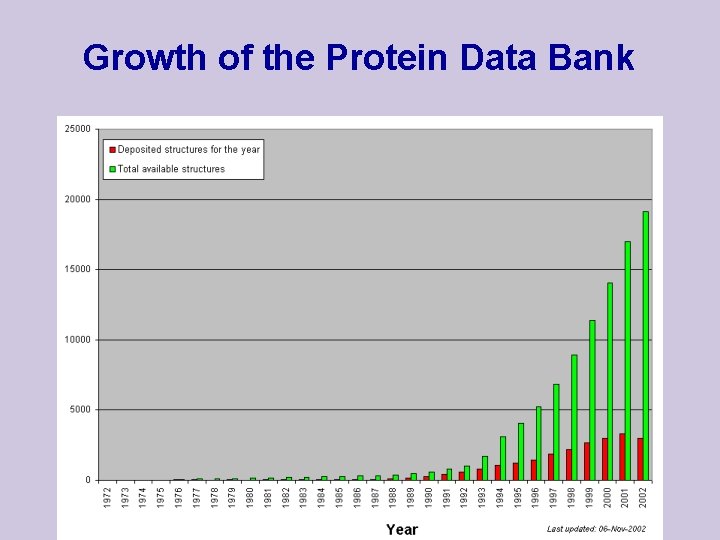
Growth of the Protein Data Bank

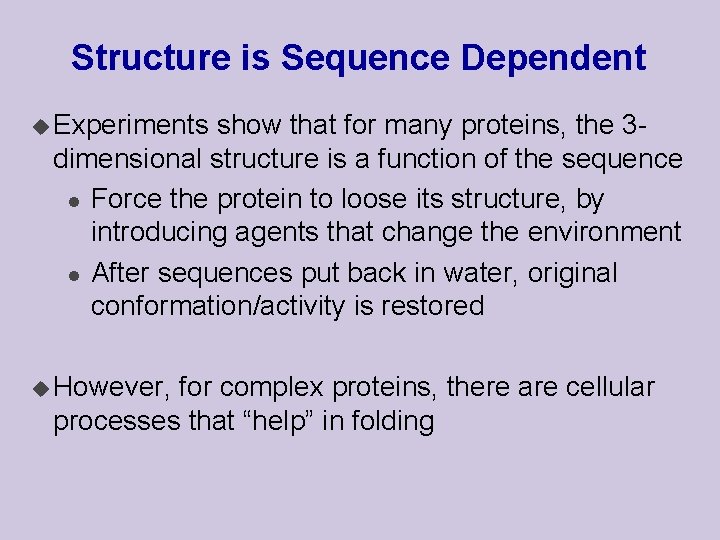
Structure is Sequence Dependent u Experiments show that for many proteins, the 3 dimensional structure is a function of the sequence l Force the protein to loose its structure, by introducing agents that change the environment l After sequences put back in water, original conformation/activity is restored u However, for complex proteins, there are cellular processes that “help” in folding

Amino Acids

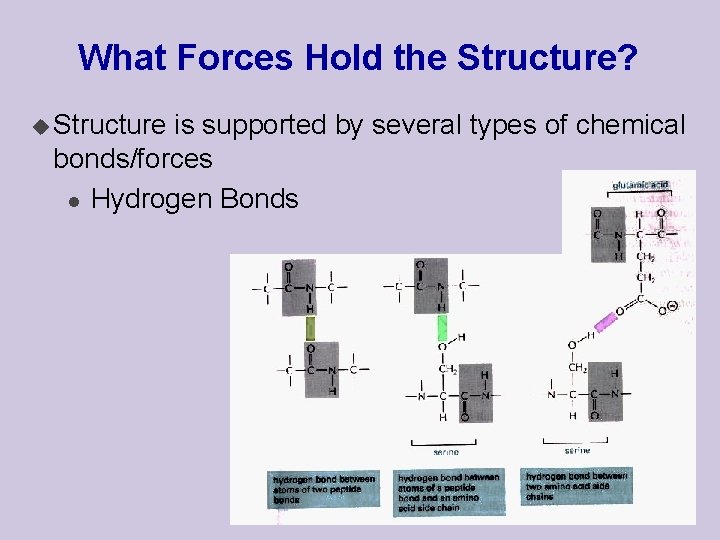
What Forces Hold the Structure? u Structure is supported by several types of chemical bonds/forces l Hydrogen Bonds

What Forces Hold the Structure? u Charge-charge l interactions Positive charged groups prefer to be situated against negatively charged groups

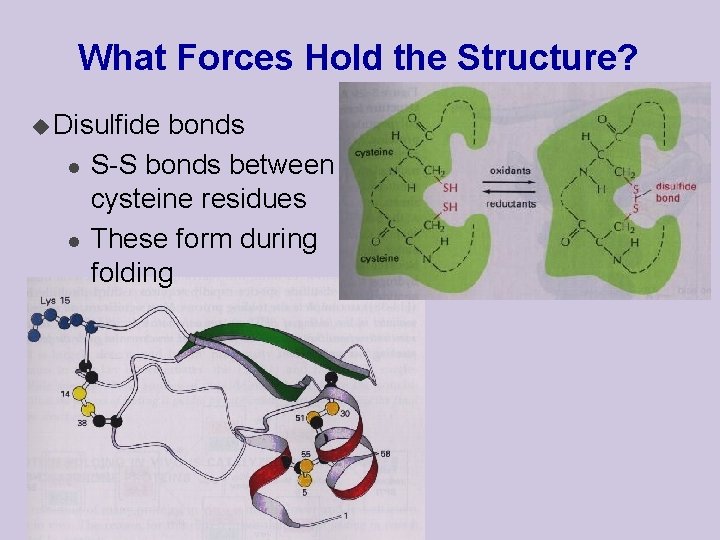
What Forces Hold the Structure? u Disulfide l l bonds S-S bonds between cysteine residues These form during folding

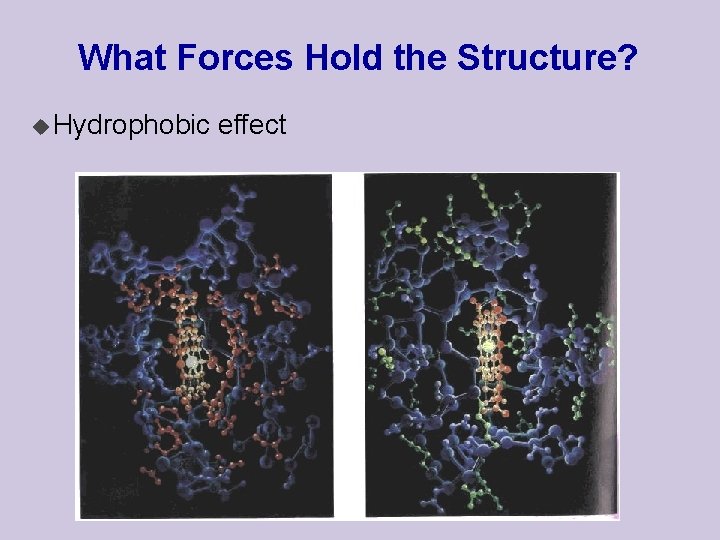
What Forces Hold the Structure? u Hydrophobic effect

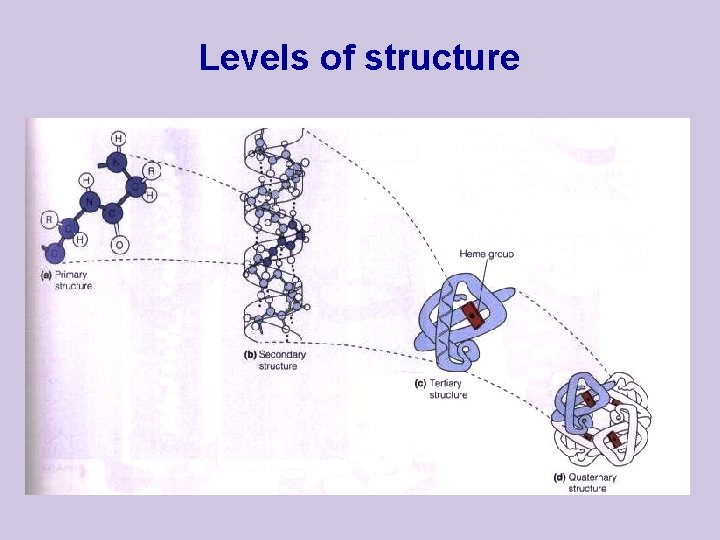
Levels of structure

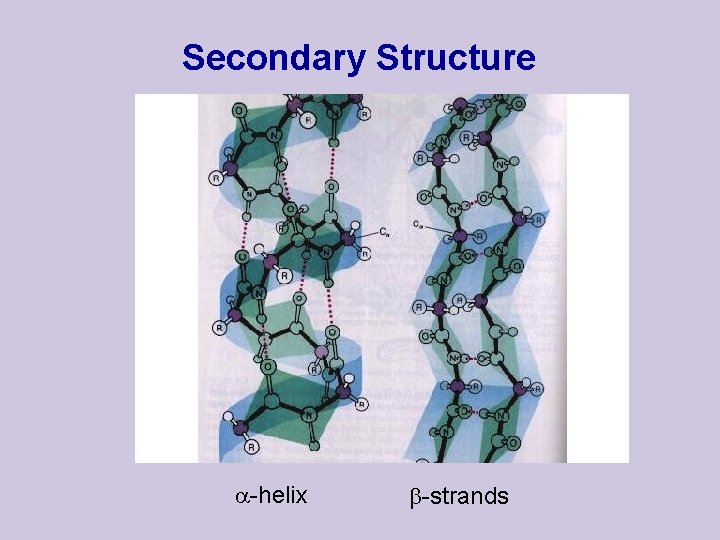
Secondary Structure -helix -strands

Hydrogen Bonds in -Helixes

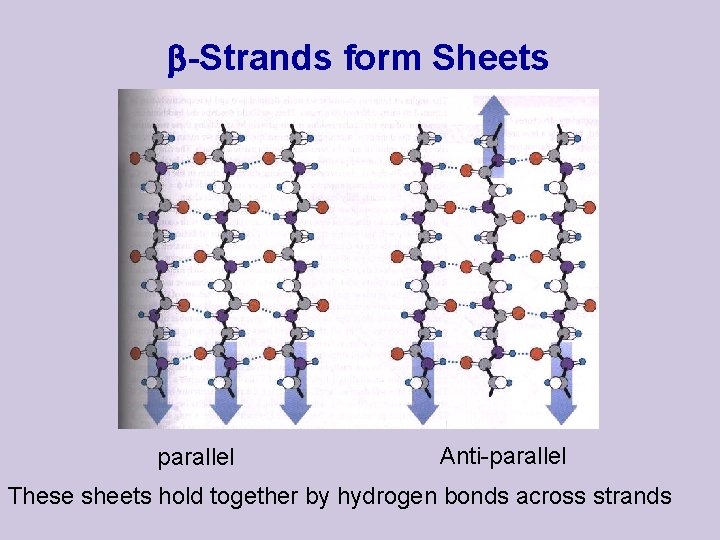
-Strands form Sheets parallel Anti-parallel These sheets hold together by hydrogen bonds across strands

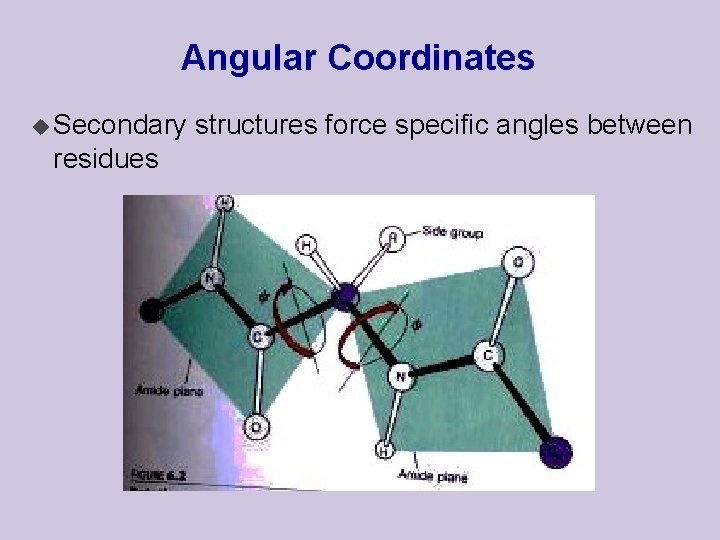
Angular Coordinates u Secondary residues structures force specific angles between

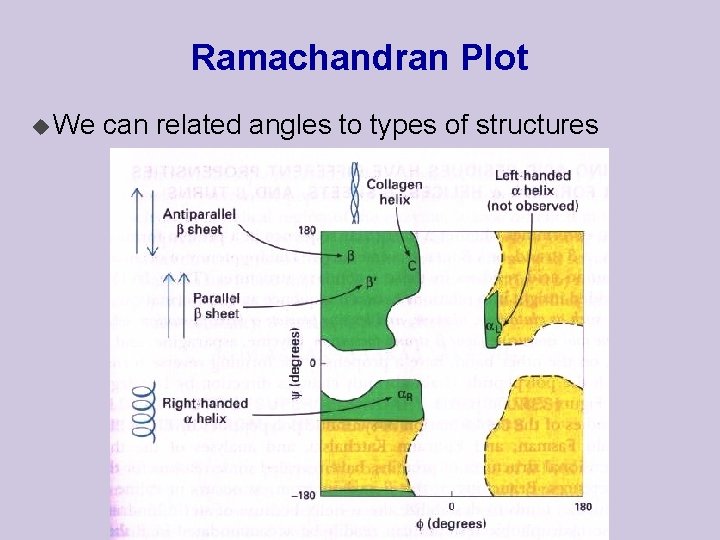
Ramachandran Plot u We can related angles to types of structures

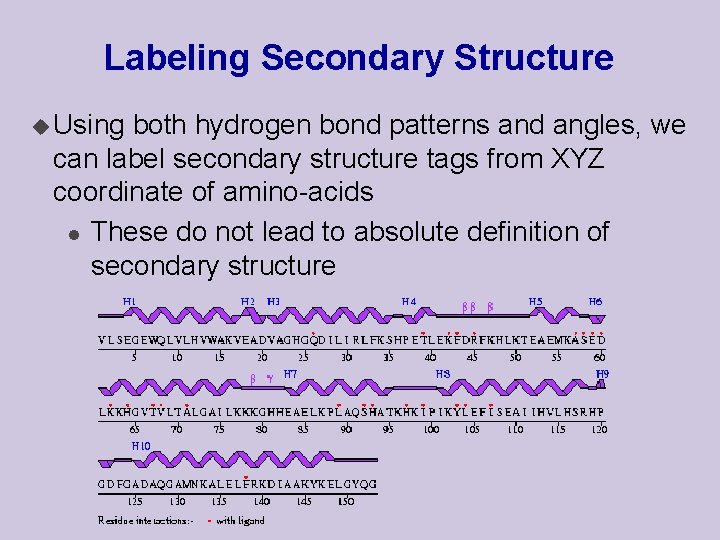
Labeling Secondary Structure u Using both hydrogen bond patterns and angles, we can label secondary structure tags from XYZ coordinate of amino-acids l These do not lead to absolute definition of secondary structure

Prediction of Secondary Structure Input: u amino-acid sequence Output: u Annotation sequence of three classes: l alpha l beta l other (sometimes called coil/turn) Measure of success: u Percentage of residues that were correctly labeled

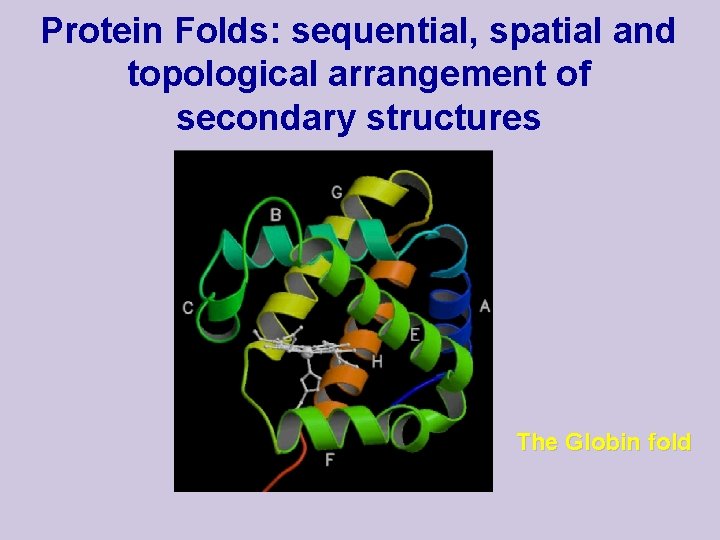
Protein Folds: sequential, spatial and topological arrangement of secondary structures The Globin fold

Approaches for structure prediction Homology modeling l (25 -30% identity as a predictor) Fold recognition l Remote homology Ab initio Prediction l Heavy computations

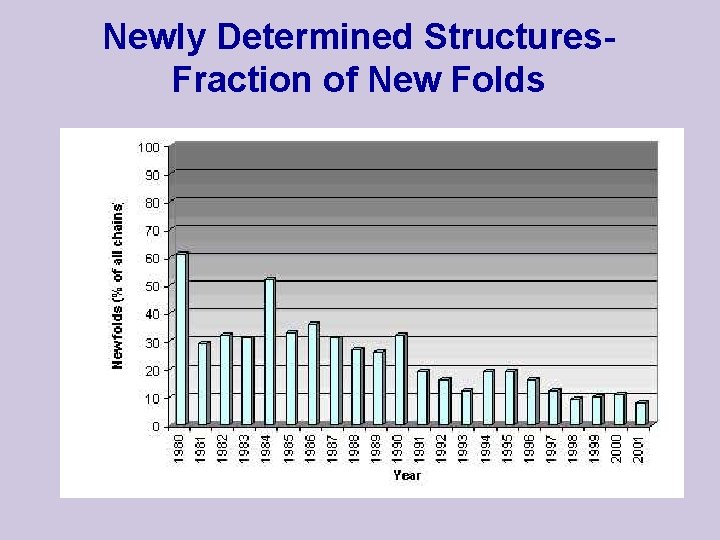
Newly Determined Structures. Fraction of New Folds

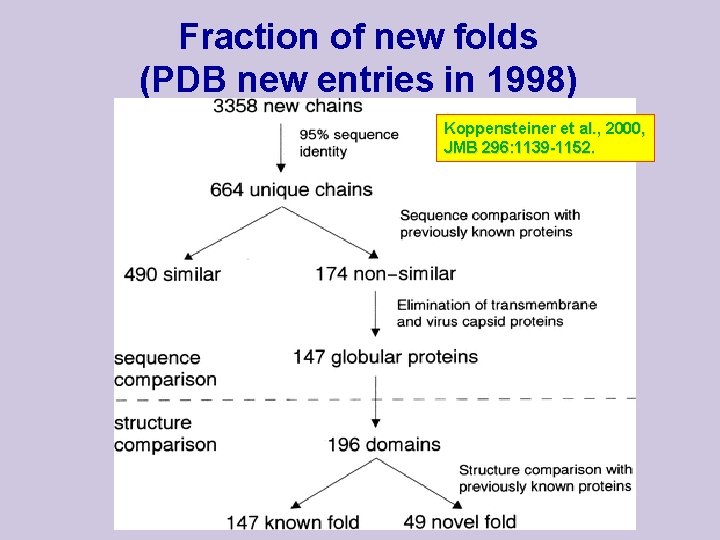
Fraction of new folds (PDB new entries in 1998) Koppensteiner et al. , 2000, JMB 296: 1139 -1152.

A Finite Number of Protein Folds Aim: recognize fold that “matches” a given sequence Approaches: l PSI-Blast, Profile HMMs, etc. l Threading

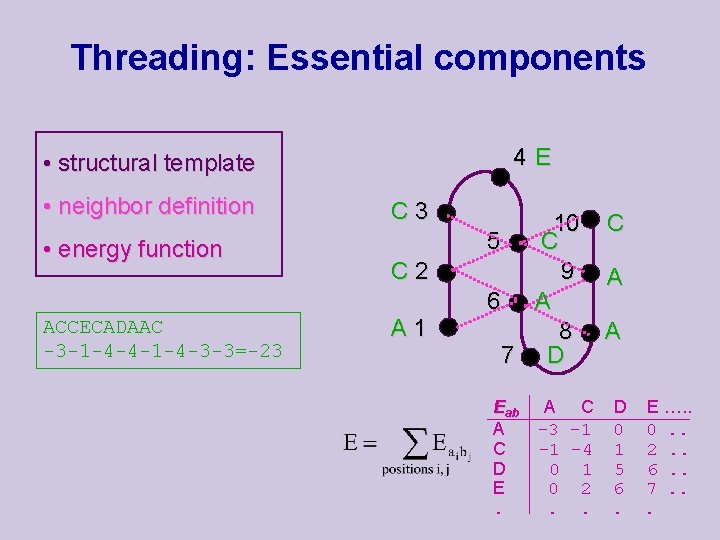
Threading: Essential components 4 E • structural template • neighbor definition • energy function ACCECADAAC -3 -1 -4 -4 -1 -4 -3 -3=-23 C 2 A 1 10 5 C 9 6 A 8 7 D Eab A C D E. A C -3 -1 -1 -4 0 1 0 2. . C A A D E …. . 0 0. . 1 2. . 5 6. . 6 7. .

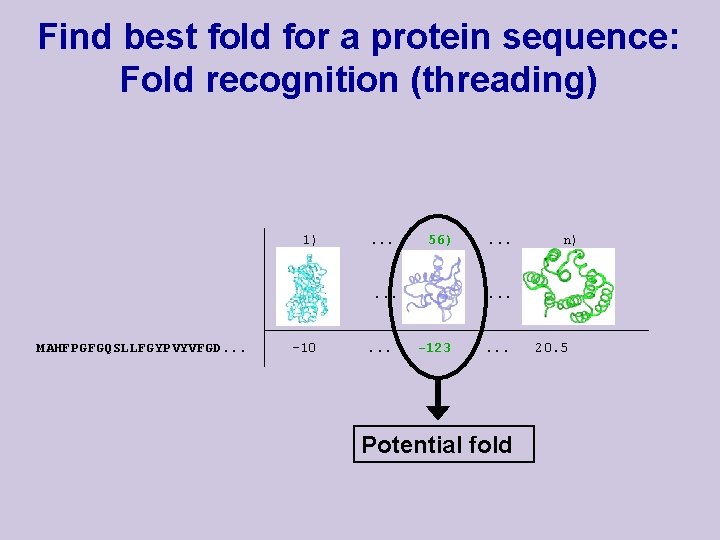
Find best fold for a protein sequence: Fold recognition (threading) 1) . . . 56) . . . MAHFPGFGQSLLFGYPVYVFGD. . . -10 . . . n) . . . -123 . . . Potential fold 20. 5

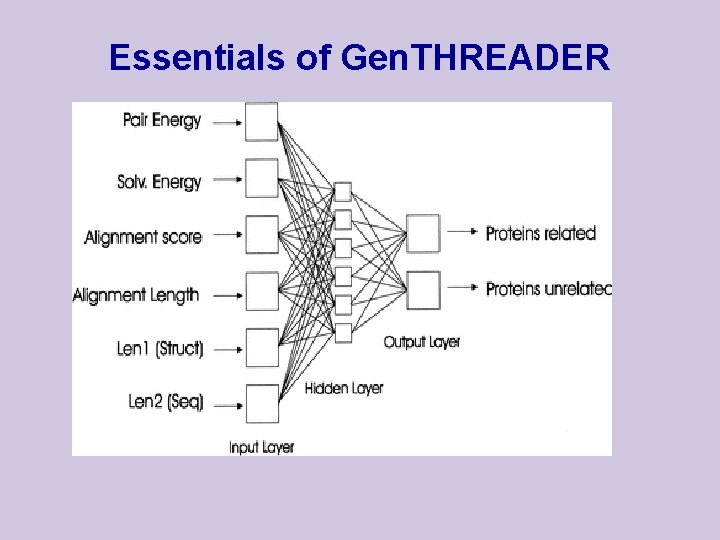
Gen. THREADER (Jones , 1999, JMB 287: 797 -815) For each template provide MSA l align the query sequence with the MSA l assess the alignment by sequence alignment score l assess the alignment by pairwise potentials l assess the alignment by solvation function l record lengths of: alignment, query, template

Essentials of Gen. THREADER

Ab-initio Structure Recognition Goal: l Predict structure from “first principles” Benefits: l Works for novel folds l Shows that we understand the process

Approaches to Ab-initio Prediction Molecular Dynamics u Simulates the forces that governs the protein within water u Since proteins natural fold, this would lead to solved structure Problems: u Thousands of atoms u Huge number of time steps to reach folded protein Intractable problem

Approaches to Ab-initio Prediction Minimal Energy u Assumption: folded form is the minimal energy conformation of the protein Decomposition: u Define energy function u Search for 3 -D conformation that minimize energy

Energy Function u Account l l l for the forces that apply on the molecule Van der wals forces Covalent bonds Hydrogen bonds Charges Hydrophobic effects Issues: u Estimating parameters u How do we compute it --- O( (# atoms)^2 )

Simplified Energy Functions Different levels of granularity u Residue-Residue energy function (Bead model) u Partial l l model Backbone as a bid Side-chain as a rigid body that can move wrt to backbone u Many other variants

Search Strategy u High dimensional search problem How do we represent partial solutions? u Position of each atom (too detailed!) u Position of each reside (too coarse!) u Intermediate solutions (e. g. , backbone and side chain)

Search Strategy Representation tradeoffs u X, Y, Z l l coordinates Easy to compute distances between residues Might represent infeasible solutions u Angles l l between successive residues Easy to ensure a “legal” protein Harder to compute distances

Search Strategy Typical approach: u Secondary structure prediction u Attempts at different conformation keeping secondary structure fixed u Finer moves relaxing secondary structure Use u Greedy search u Simulated annealing u…

Rosetta Method Idea: l “Structural” signatures are reoccurring within protein structures l Use these as cues during structure search

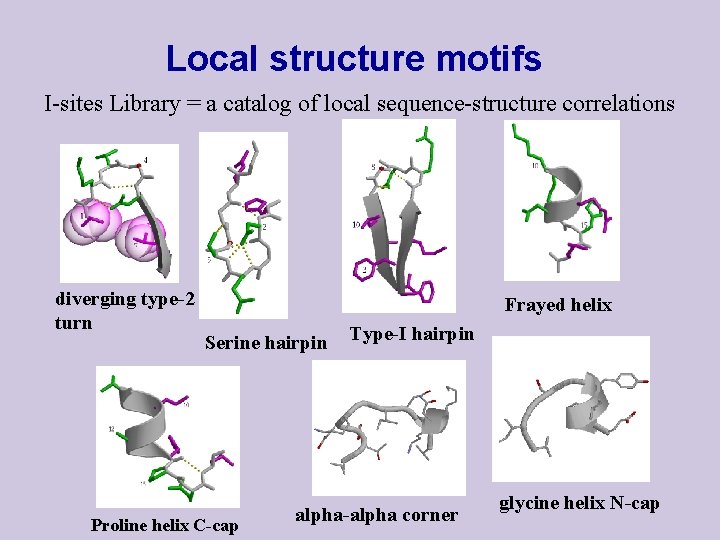
Local structure motifs I-sites Library = a catalog of local sequence-structure correlations diverging type-2 turn Frayed helix Serine hairpin Proline helix C-cap Type-I hairpin alpha-alpha corner glycine helix N-cap

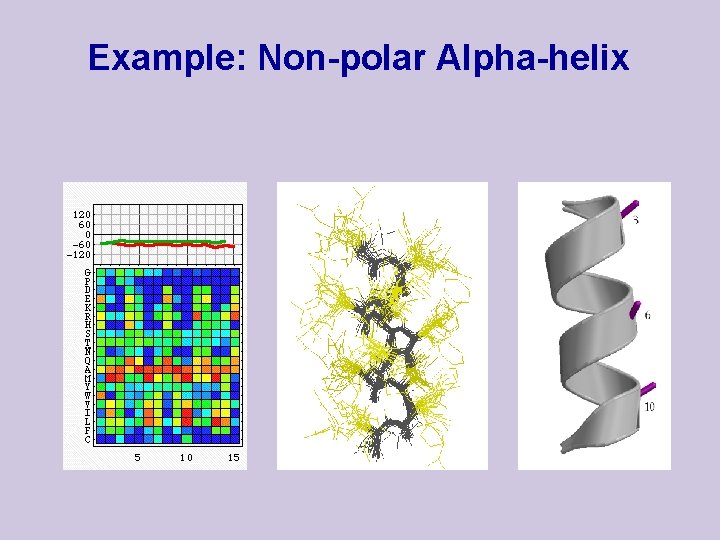
Example: Non-polar Alpha-helix

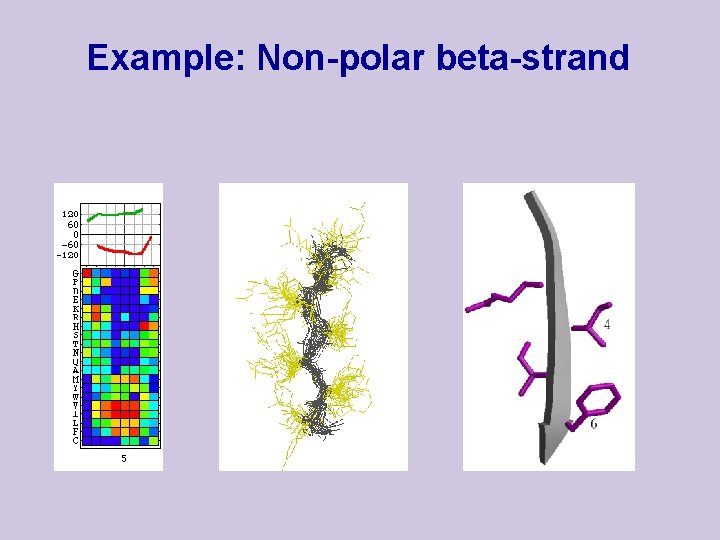
Example: Non-polar beta-strand

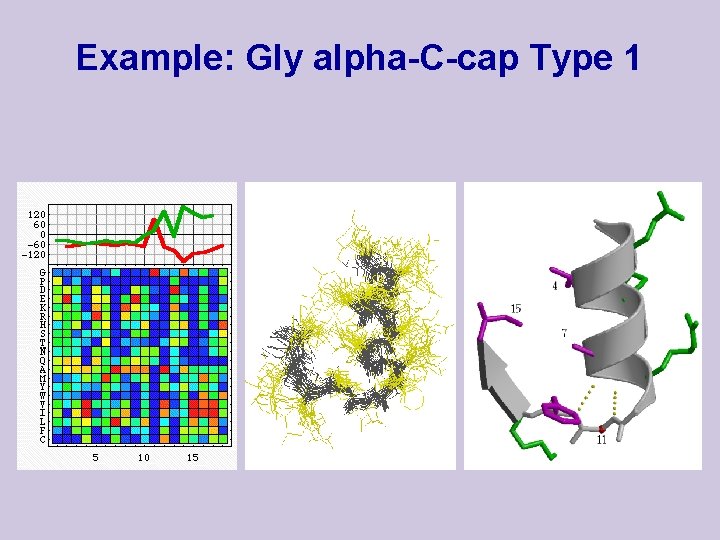
Example: Gly alpha-C-cap Type 1

Construction of I-sites library u Construct profiles (PSI-BLAST like) for each solved structure u Collect each possible segments of fixed length (len = 3, 9, 15) u Perform k-means clustering of segments u Check each cluster for a “coherent” structure (in terms of dihedral angles u Prune incoherent structures u Iteratively refine remaining clusters by removing structurally different segments, redefining cluster membership, etc.

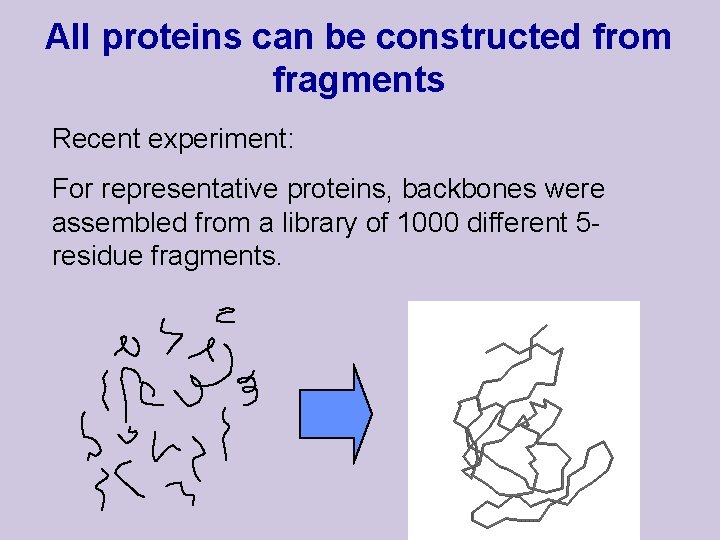
All proteins can be constructed from fragments Recent experiment: For representative proteins, backbones were assembled from a library of 1000 different 5 residue fragments.

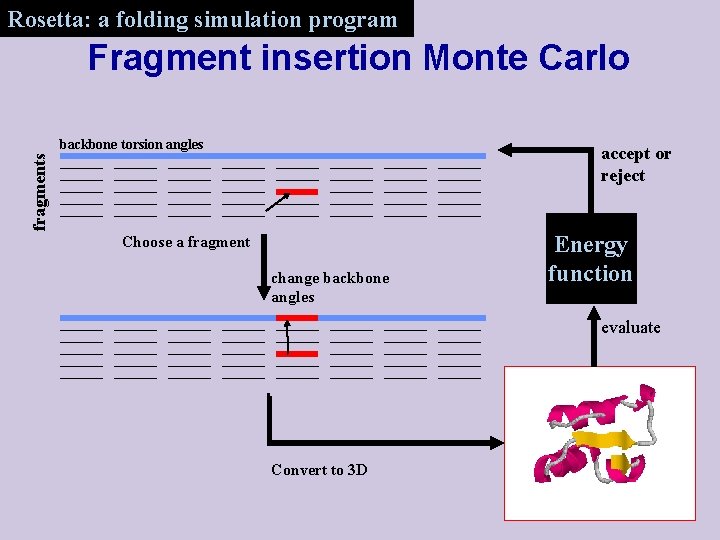
Rosetta: a folding simulation program Fragment insertion Monte Carlo backbone torsion angles fragments accept or reject Choose a fragment change backbone angles Energy function evaluate Convert to 3 D

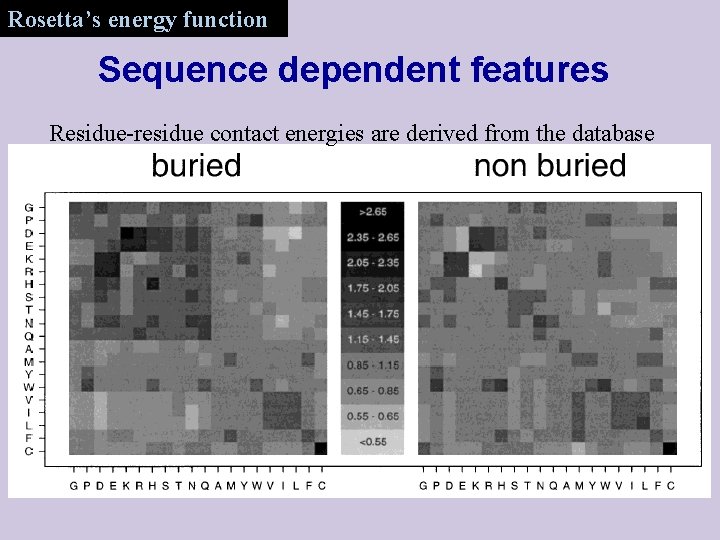
Rosetta’s energy function Sequence dependent features Residue-residue contact energies are derived from the database

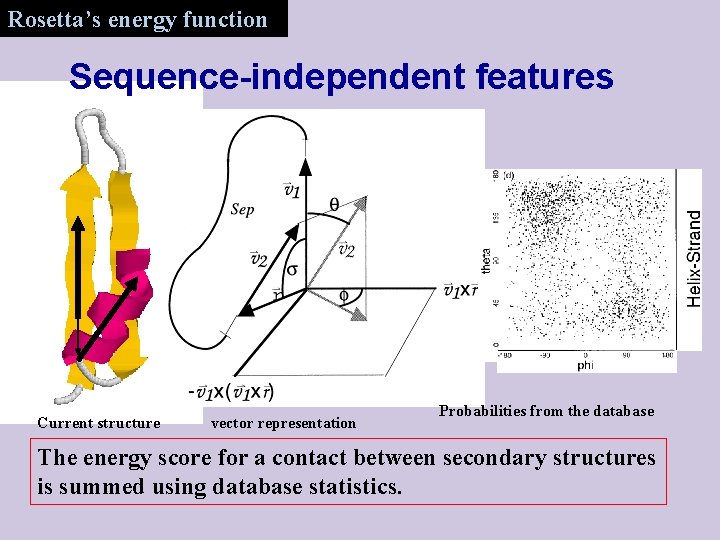
Rosetta’s energy function Sequence-independent features Current structure vector representation Probabilities from the database The energy score for a contact between secondary structures is summed using database statistics.

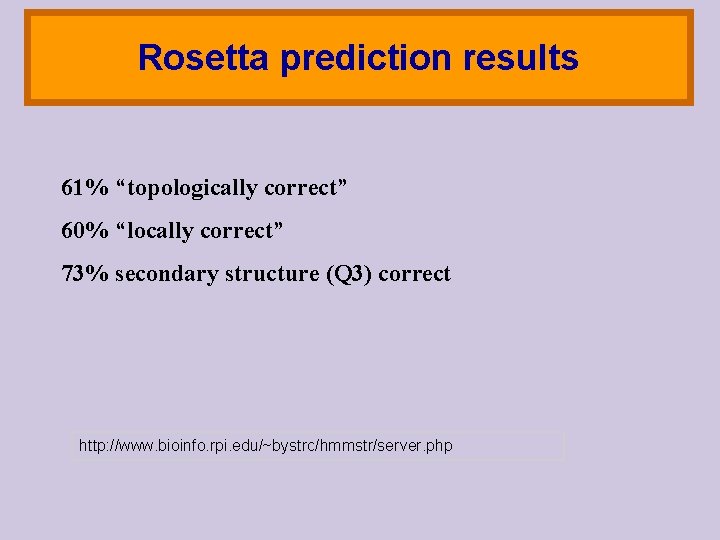
Rosetta prediction results 61% “topologically correct” 60% “locally correct” 73% secondary structure (Q 3) correct http: //www. bioinfo. rpi. edu/~bystrc/hmmstr/server. php

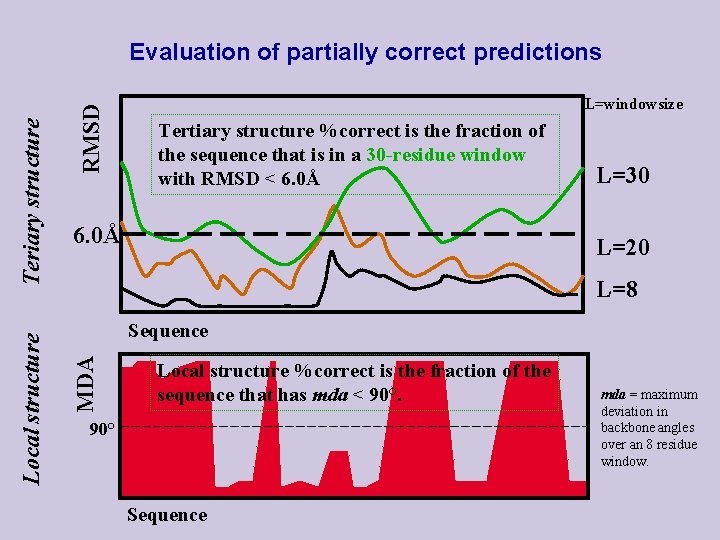
RMSD L=windowsize Tertiary structure %correct is the fraction of the sequence that is in a 30 -residue window with RMSD < 6. 0Å L=30 L=20 L=8 Sequence MDA Local structure Teriary structure Evaluation of partially correct predictions Local structure %correct is the fraction of the sequence that has mda < 90° Sequence mda = maximum deviation in backbone angles over an 8 residue window.

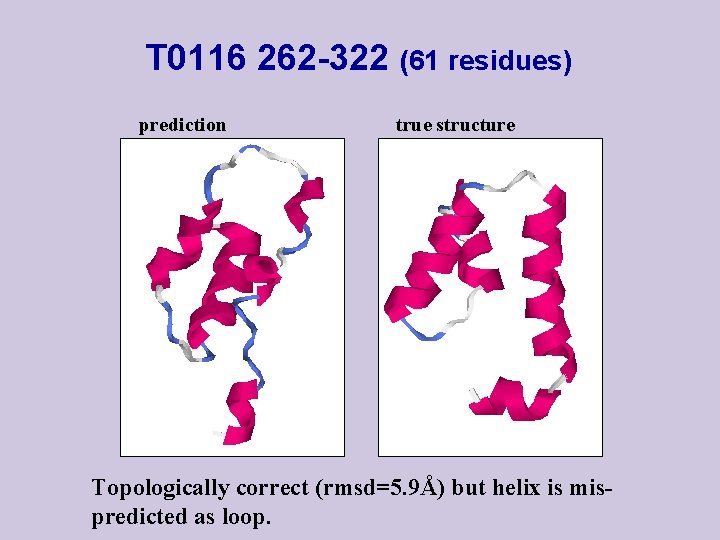
T 0116 262 -322 (61 residues) prediction true structure Topologically correct (rmsd=5. 9Å) but helix is mispredicted as loop.

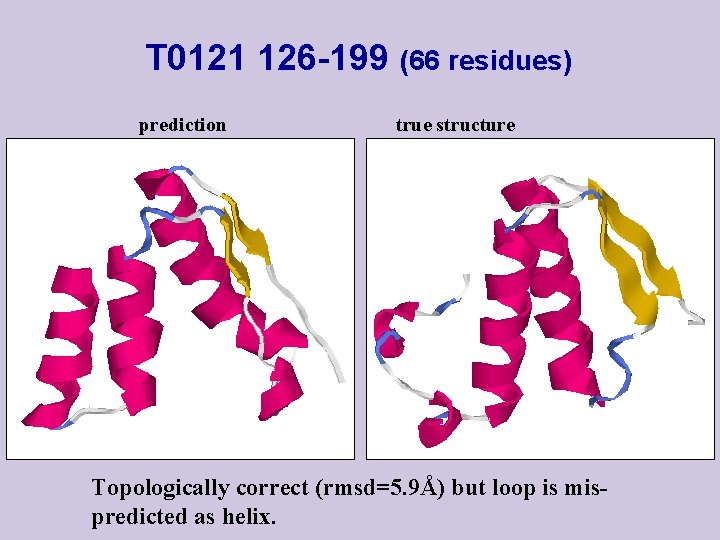
T 0121 126 -199 (66 residues) prediction true structure Topologically correct (rmsd=5. 9Å) but loop is mispredicted as helix.

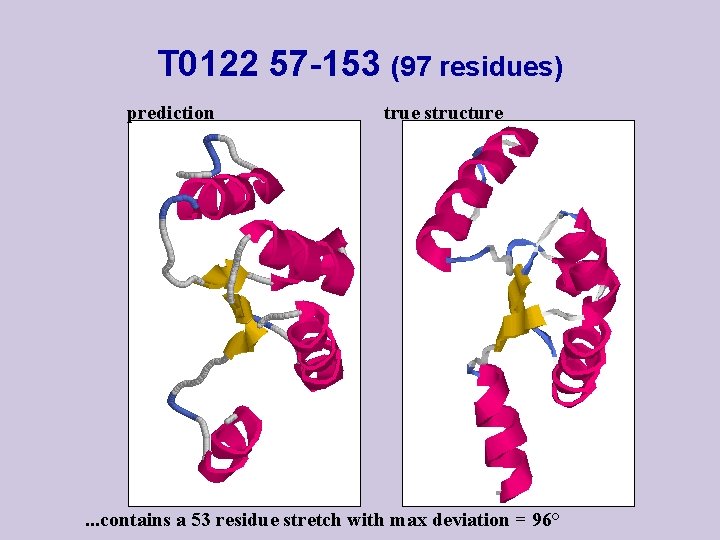
T 0122 57 -153 (97 residues) prediction true structure . . . contains a 53 residue stretch with max deviation = 96°

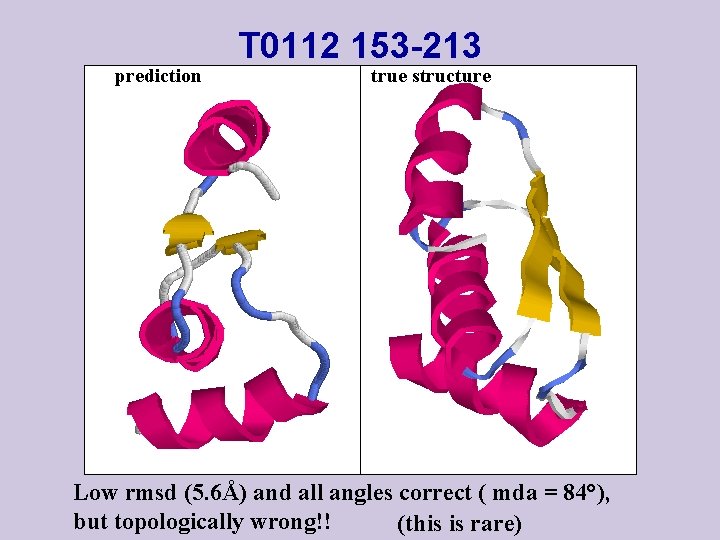
prediction T 0112 153 -213 true structure Low rmsd (5. 6Å) and all angles correct ( mda = 84°), but topologically wrong!! (this is rare)