Protein Structure Prediction II SCOP CATH gen THREADER


- Slides: 25

Protein Structure Prediction II • • • SCOP CATH gen. THREADER Swiss-Model Mod. Base – – - Protein structure classification 3 D structure prediction A database of 3 D struc. Predict.

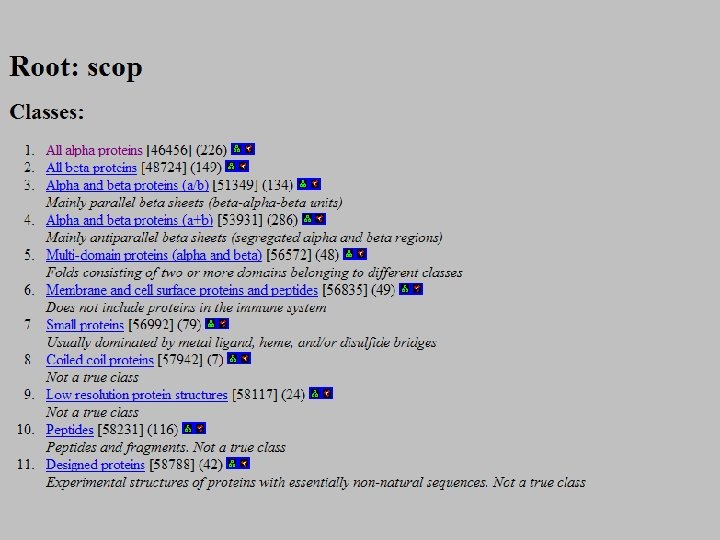
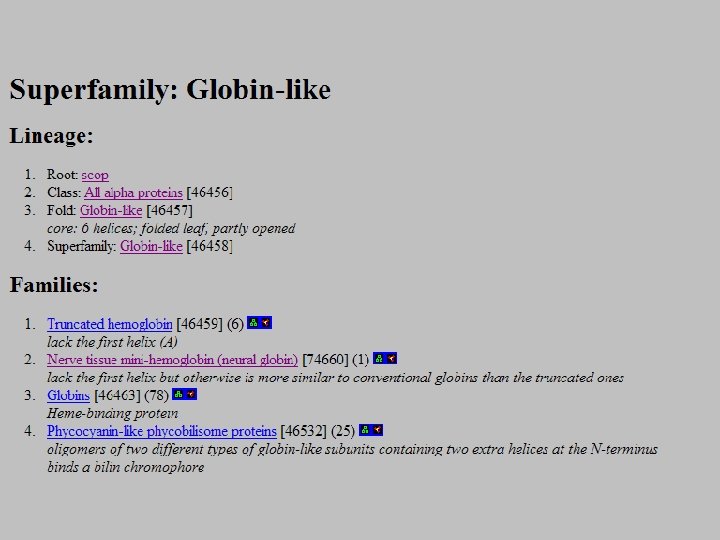
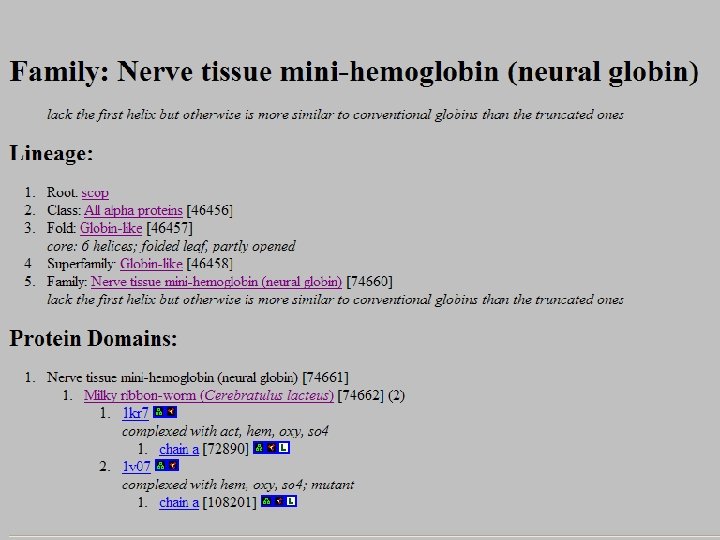
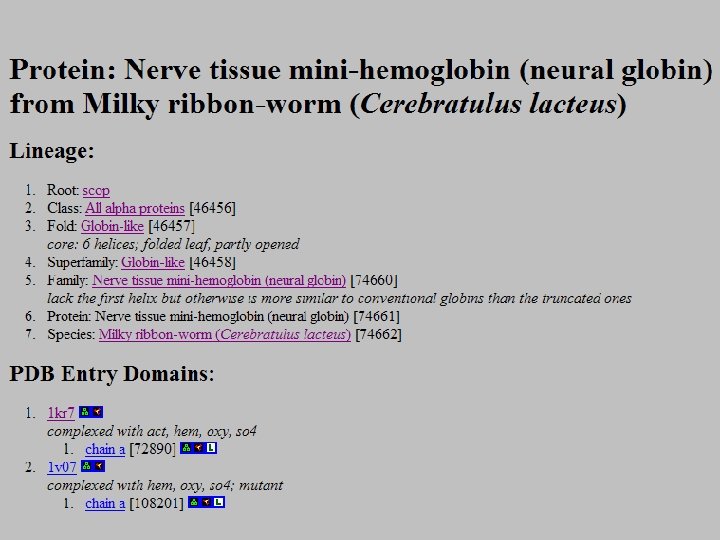
SCOP: Structural Classification of Proteins http: //scop. mrc-lmb. cam. ac. uk/scop/ • Based on known protein structures • Manually created by visual inspection • Hierarchical database structure: –Class, Fold, Superfamily, Family, Protein and Species


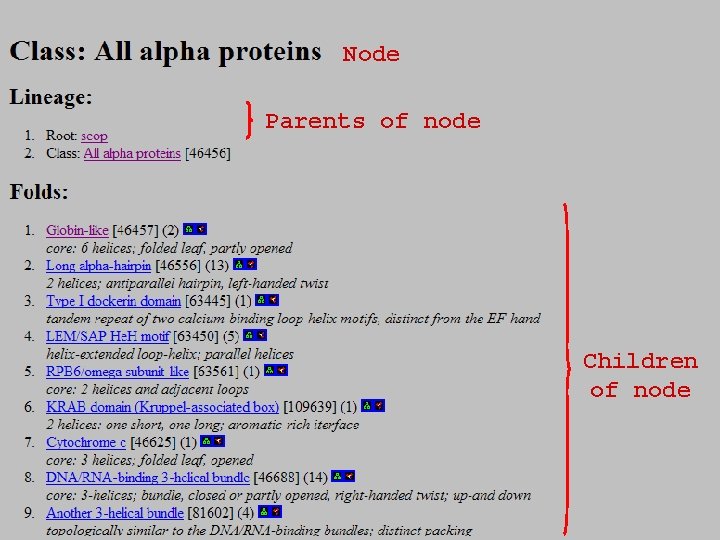
Node Parents of node Children of node

Node Parents of node Children of node




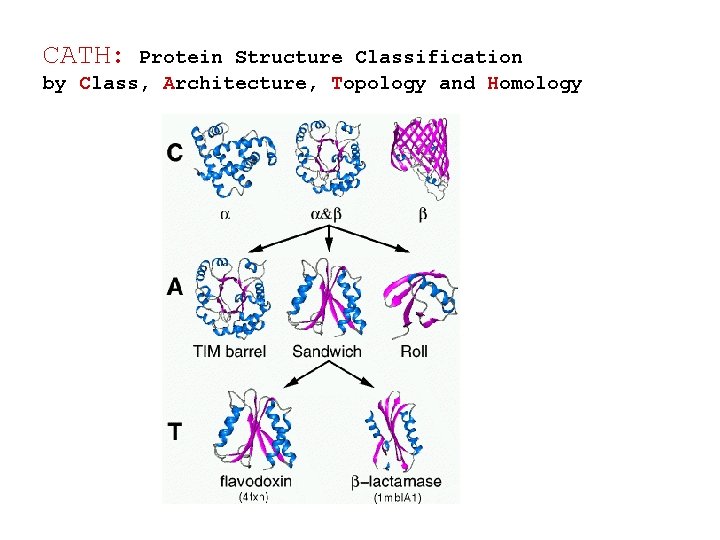
CATH: Protein Structure Classification by Class, Architecture, Topology and Homology http: //www. cathdb. info/latest/index. html • Class: The secondary structure composition: mainly-alpha, mainly-beta and alpha-beta. • Architecture: The overall shape of the domain structure. Orientations of the secondary structures : e. g. barrel or 3 layer sandwich. • Topology: Structures are grouped into fold groups at this level depending on both the overall shape and connectivity of the secondary structures. • Homologous Superfamily: Evolutionary conserved structures

CATH: Protein Structure Classification by Class, Architecture, Topology and Homology


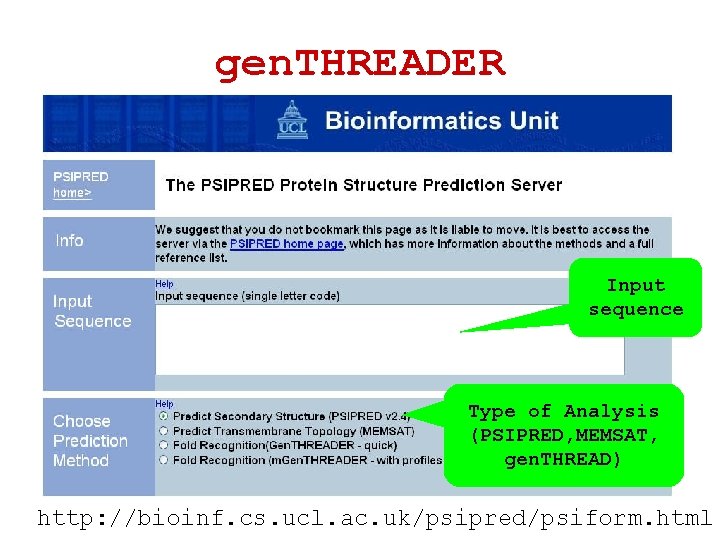
gen. THREADER Input sequence Type of Analysis (PSIPRED, MEMSAT, gen. THREAD) http: //bioinf. cs. ucl. ac. uk/psipred/psiform. html

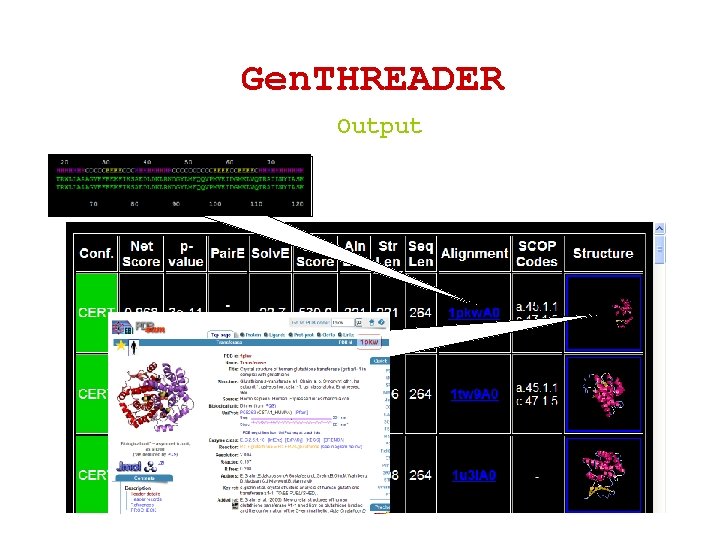
Gen. THREADER Output

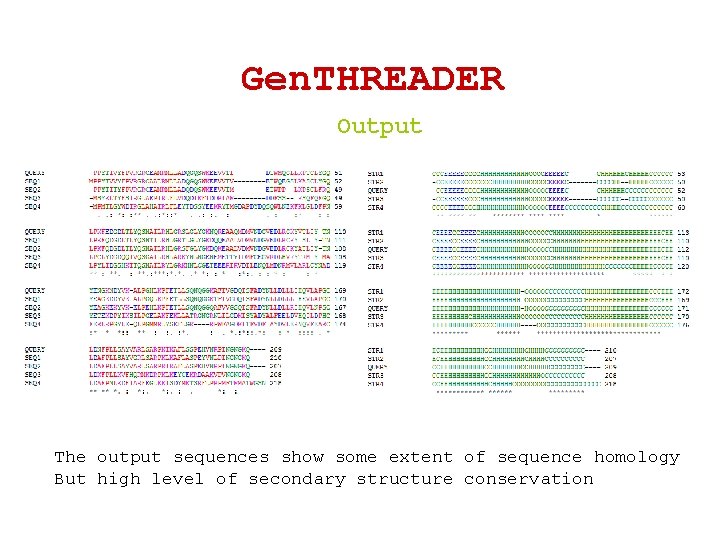
Gen. THREADER Output The output sequences show some extent of sequence homology But high level of secondary structure conservation

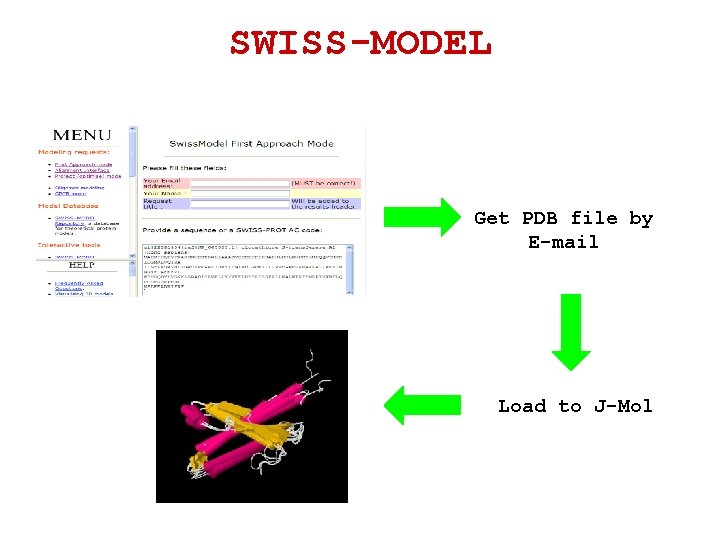
SWISS-MODEL An automated protein modeling server. http: //swissmodel. expasy. org/

SWISS-MODEL • The SWISS-MODEL algorithm can be divided into three steps: 1. Search for suitable templates: the server finds all similarities of a query sequence to sequences of known structure. It uses the BLASTP 2 program with the Ex. NRL-3 D database (a derivative of PDB database, specified for SWISS-MODEL). You get these partial results as a Swiss. Model Trace. Log file. 2. Check sequence identity with target: All templates with sequence identities above 25% are selected 3. Create the model using the Pro. Mod. II program. You get this as a Swiss. Model-Model file.

SWISS-MODEL Get PDB file by E-mail Load to J-Mol

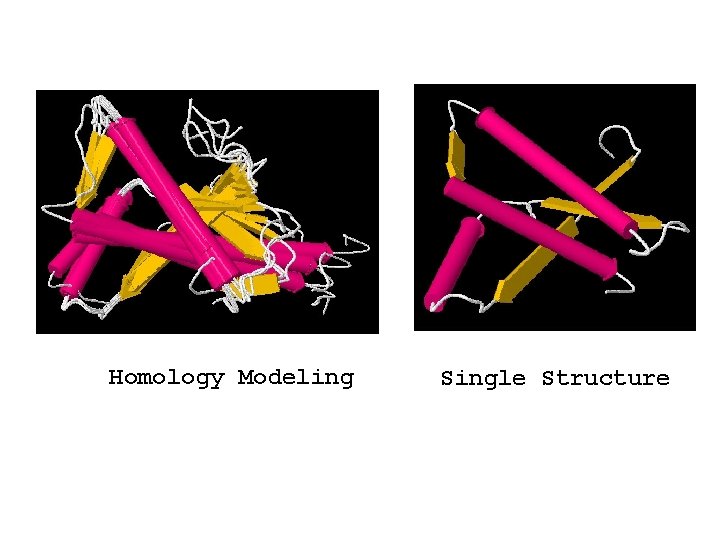
Homology Modeling Single Structure

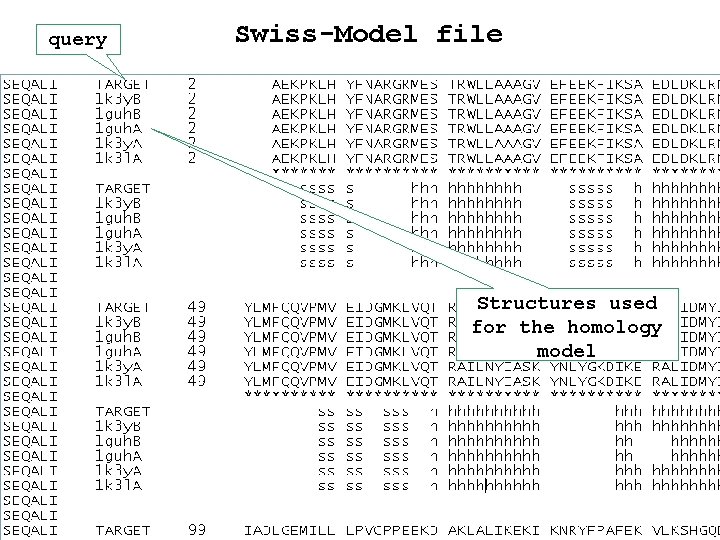
query Swiss-Model file Structures used for the homology model

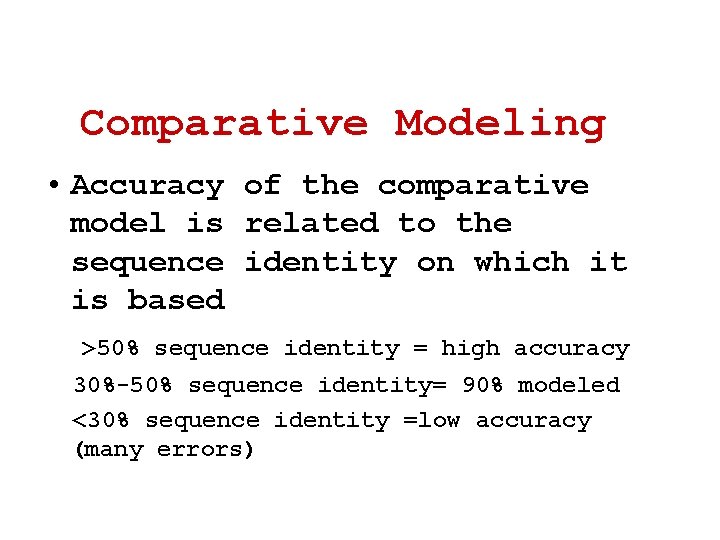
Comparative Modeling • Accuracy of the comparative model is related to the sequence identity on which it is based >50% sequence identity = high accuracy 30%-50% sequence identity= 90% modeled <30% sequence identity =low accuracy (many errors)

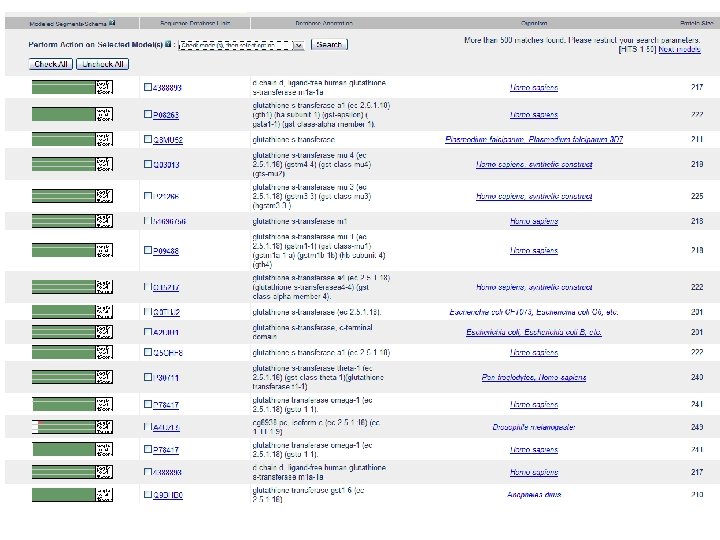
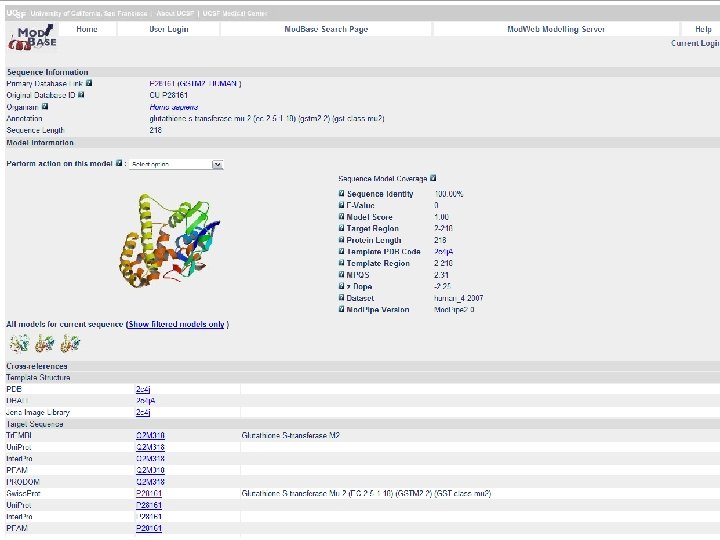
Mod. Base A Homology Model Database



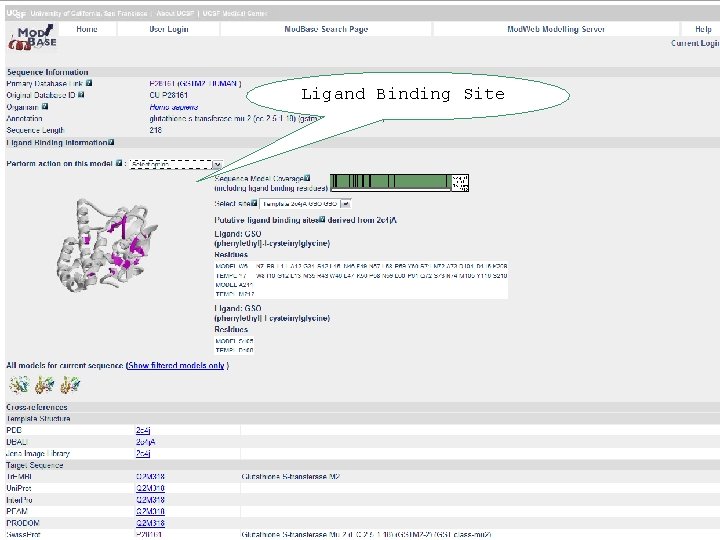
Ligand Binding Site

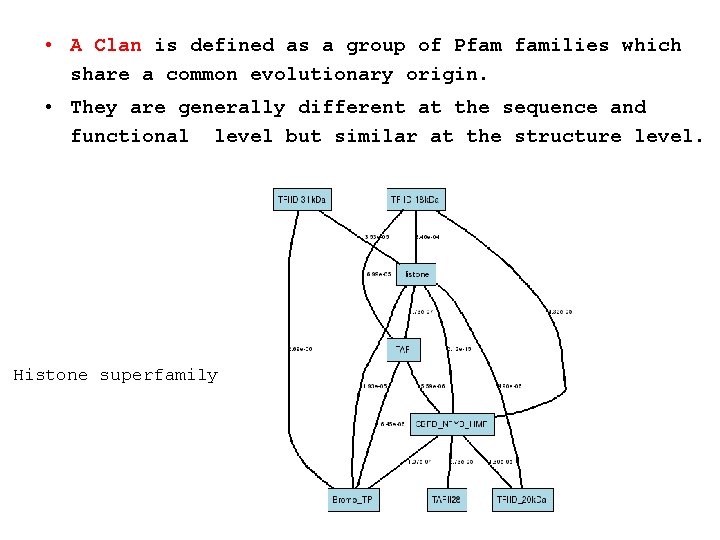
• A Clan is defined as a group of Pfam families which share a common evolutionary origin. • They are generally different at the sequence and functional level but similar at the structure level. Histone superfamily