Protein Structure Prediction and Analysis Tools Jianlin Cheng













- Slides: 65

Protein Structure Prediction and Analysis Tools Jianlin Cheng, Ph. D Assistant Professor Computer Science Department & Informatics Institute University of Missouri, Columbia 2012

Sequence, Structure and Function AGCWY…… Cell

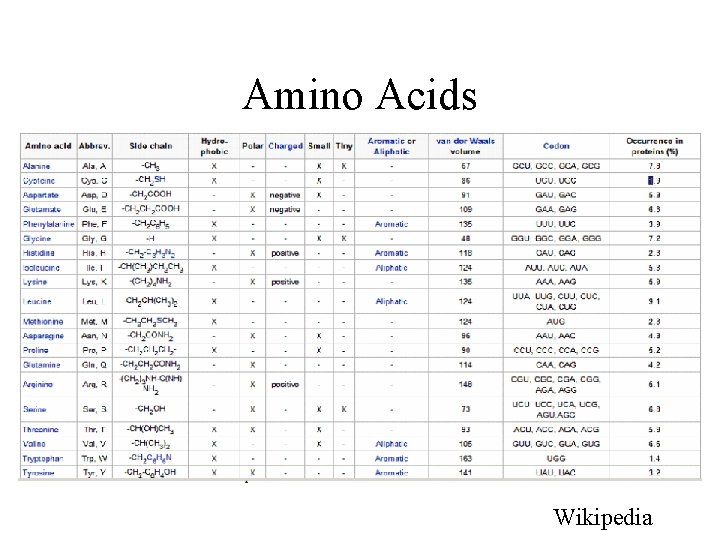
Amino Acids Wikipedia

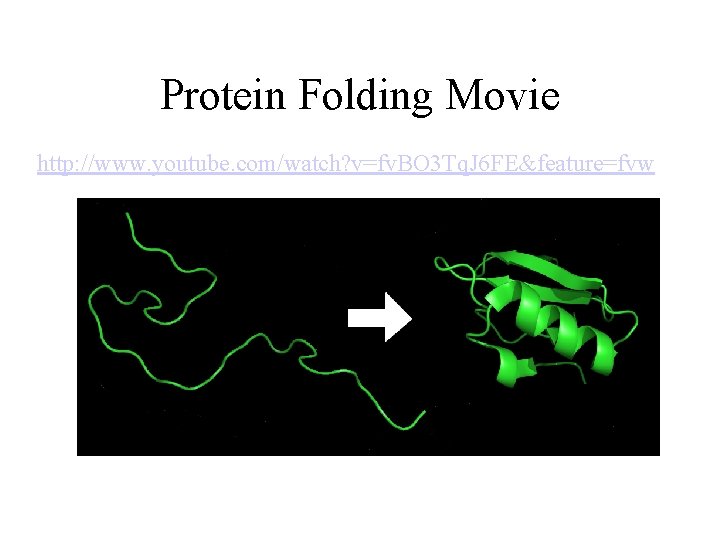
Protein Folding Movie http: //www. youtube. com/watch? v=fv. BO 3 Tq. J 6 FE&feature=fvw

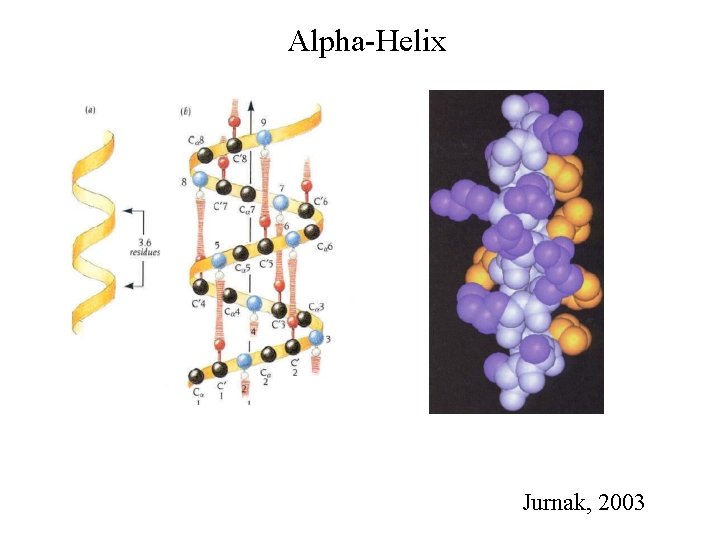
Alpha-Helix Jurnak, 2003

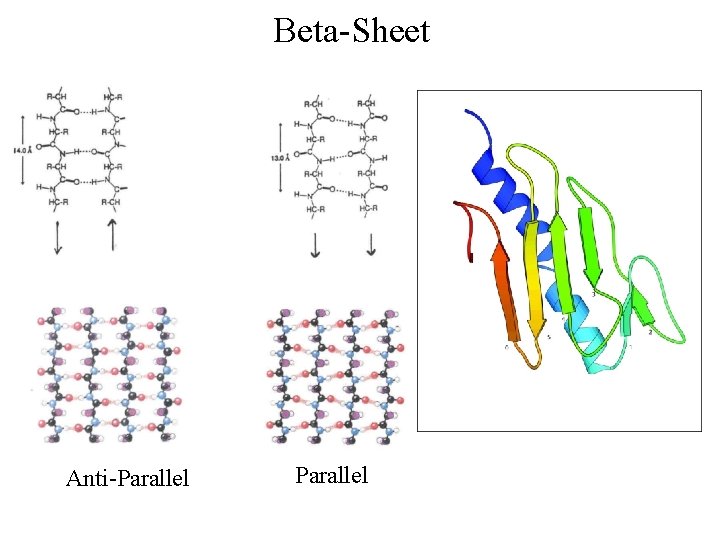
Beta-Sheet Anti-Parallel

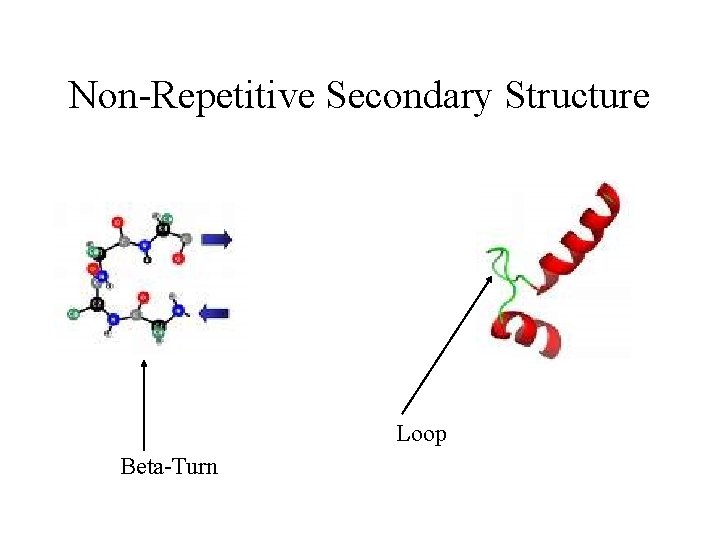
Non-Repetitive Secondary Structure Loop Beta-Turn

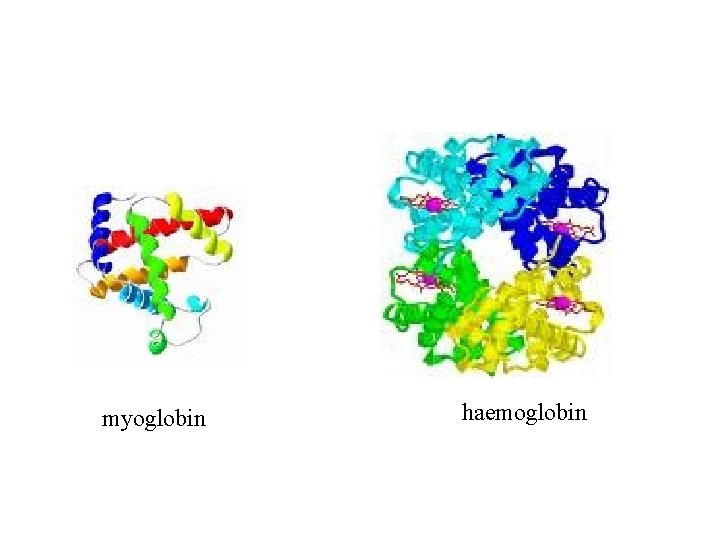
myoglobin haemoglobin

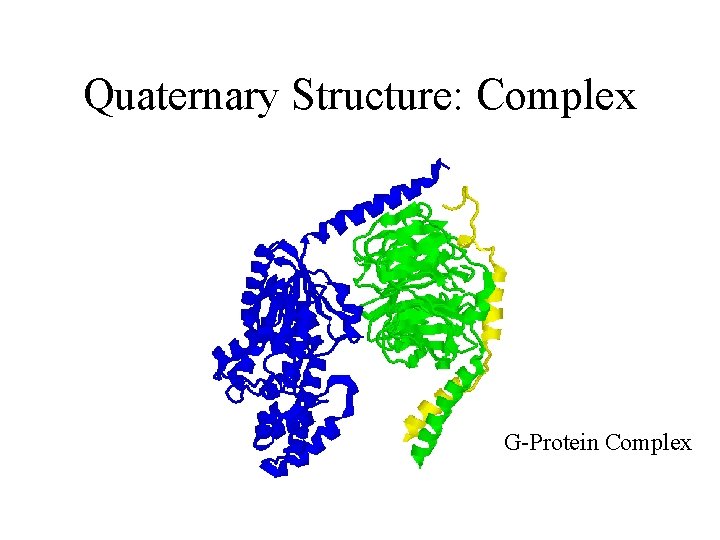
Quaternary Structure: Complex G-Protein Complex

Protein Structure Determination • X-ray crystallography • Nuclear Magnetic Resonance (NMR) Spectroscopy • X-ray: any size, accurate (1 -3 Angstrom (10 -10 m)), sometime hard to grow crystal • NMR: small to medium size, moderate accuracy, structure in solution

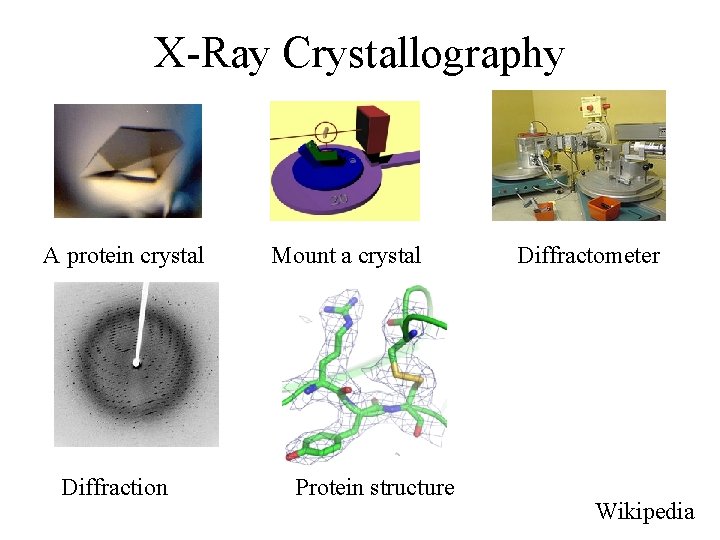
X-Ray Crystallography A protein crystal Diffraction Mount a crystal Protein structure Diffractometer Wikipedia

Pacific Northwest National Laboratory's high magnetic field (800 MHz, 18. 8 T) NMR spectrometer being loaded with a sample. Wikipedia, the free encyclopedia

Storage in Protein Data Bank Search database

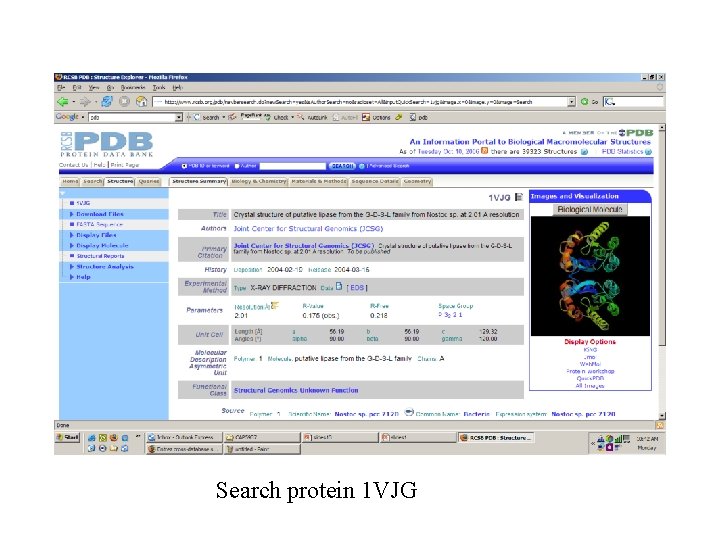
Search protein 1 VJG

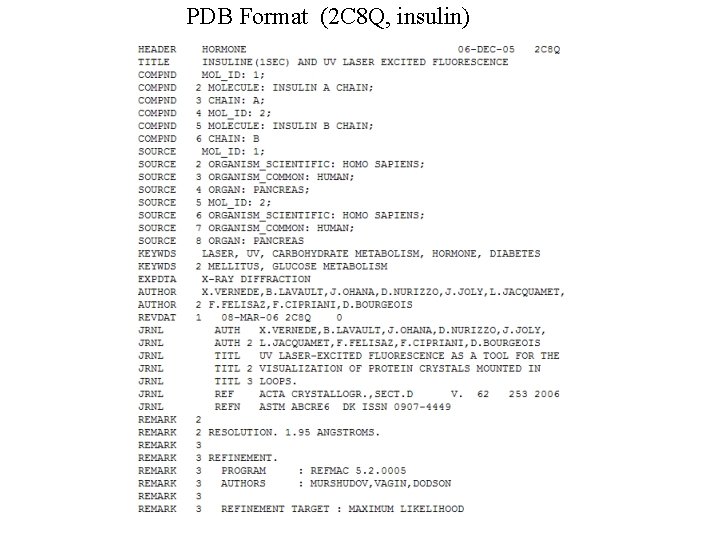
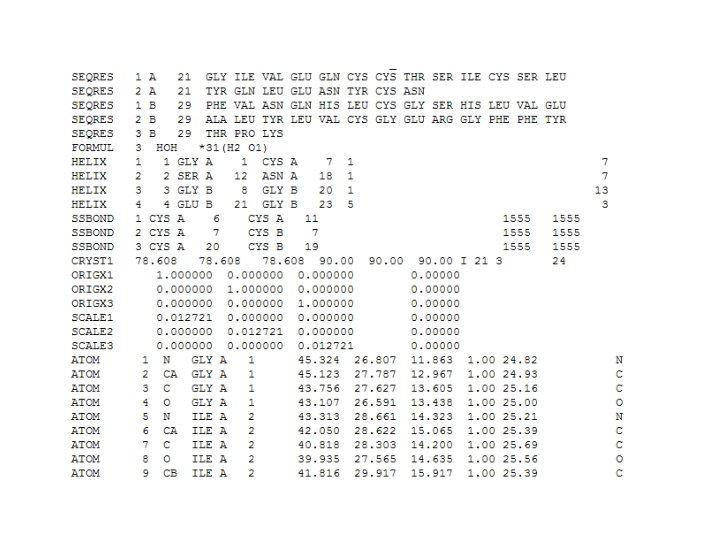
PDB Format (2 C 8 Q, insulin)


Structure Visualization • Rasmol (http: //www. umass. edu/microbio/rasmol/getras. ht m) • MDL Chime (plug-in) (http: //www. mdl. com/products/framework/chime/) • Protein Explorer (http: //molvis. sdsc. edu/protexpl/frntdoor. htm) • Jmol: http: //jmol. sourceforge. net/ • Pymol: http: //pymol. sourceforge. net/

J. Pevsner, 2005

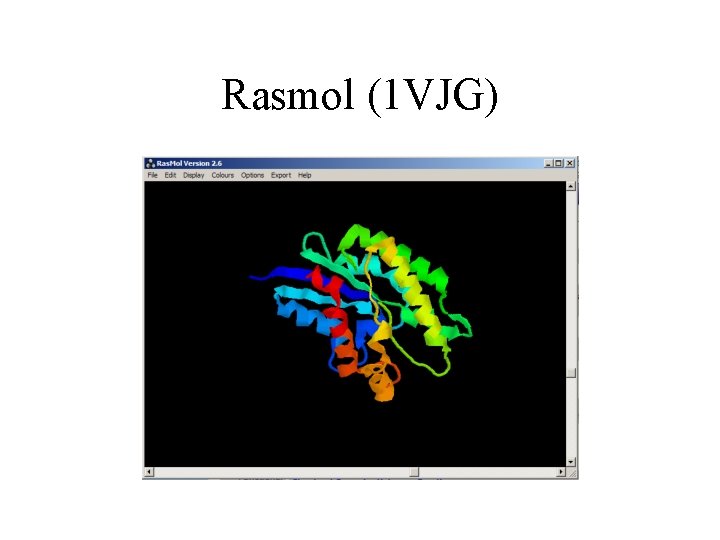
Rasmol (1 VJG)

Structure Analysis • Assign secondary structure for amino acids from 3 D structure • Generate solvent accessible area for amino acids from 3 D structure • Most widely used tool: DSSP (Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Kabsch and Sander, 1983)

DSSP server: http: //bioweb. pasteur. fr/seqanal/interfaces/dssp-simple. html DSSP download: http: //swift. cmbi. ru. nl/gv/dssp/ DSSP Code: H = alpha helix G = 3 -helix (3/10 helix) I = 5 helix (pi helix) B = residue in isolated beta-bridge E = extended strand, participates in beta ladder T = hydrogen bonded turn S = bend Blank = loop

DSSP Web Service http: //bioweb. pasteur. fr/seqanal/interfaces/dssp-simple. html

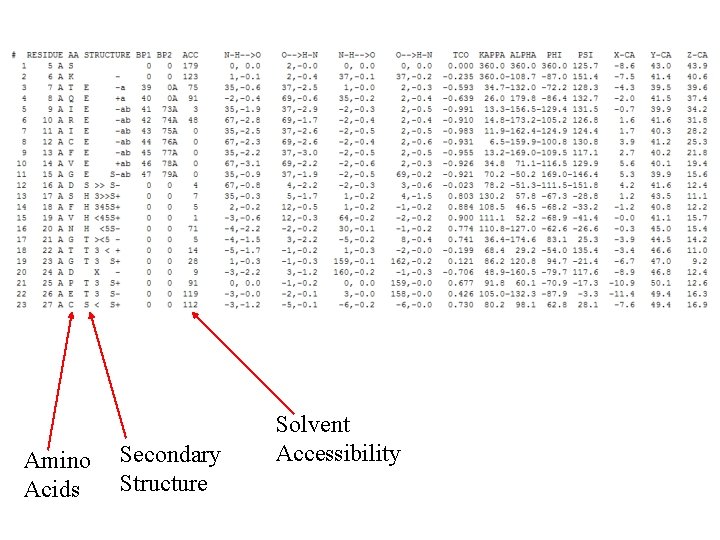
Amino Acids Secondary Structure Solvent Accessibility

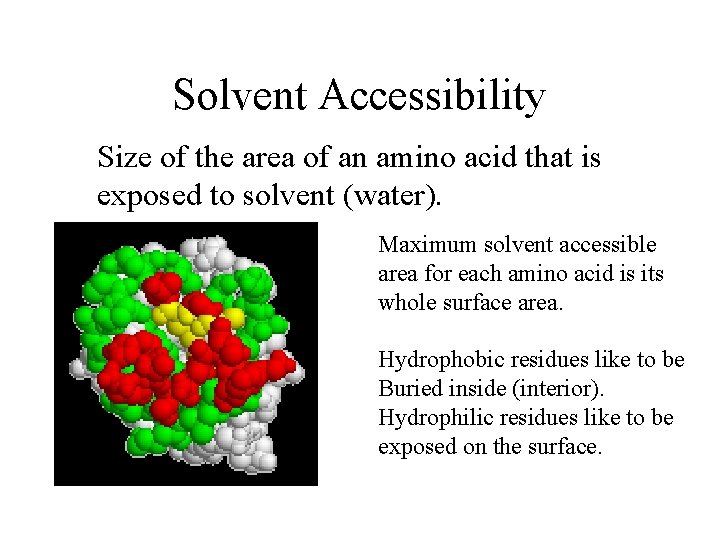
Solvent Accessibility Size of the area of an amino acid that is exposed to solvent (water). Maximum solvent accessible area for each amino acid is its whole surface area. Hydrophobic residues like to be Buried inside (interior). Hydrophilic residues like to be exposed on the surface.

Structure Comparison (Alignment) • Are the structures of two protein similar? • Are the two structure models of the same protein similar? • Different measures (RMSD, GDT-TS (Zemla et al. , 1999), Max. Sub (Siew et al. , 2000), TM score (Zhang and Skolnick, 2005))

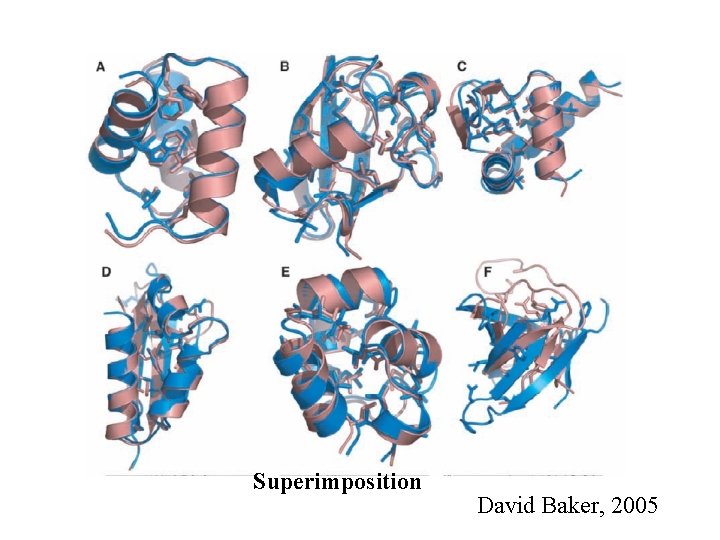
Superimposition David Baker, 2005

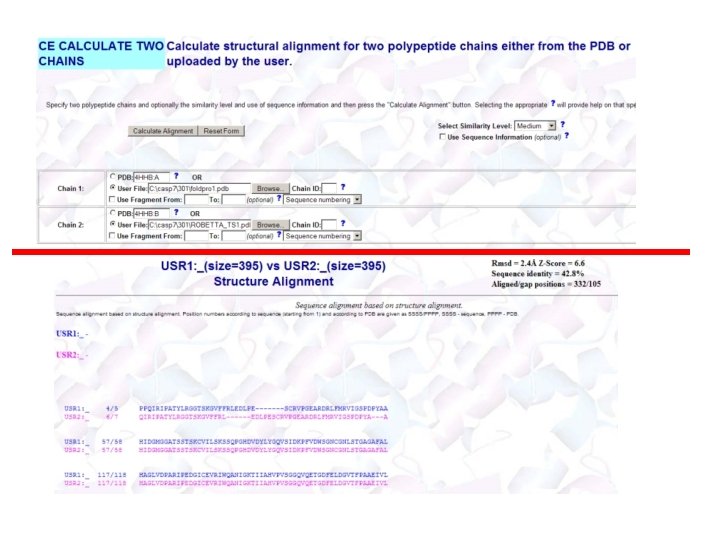
Useful Structure Alignment Tools • CE (http: //cl. sdsc. edu/) • DALI (http: //www. ebi. ac. uk/ dali/) • TM-Align: http: //zhang. bioinform atics. ku. edu/TM-align/


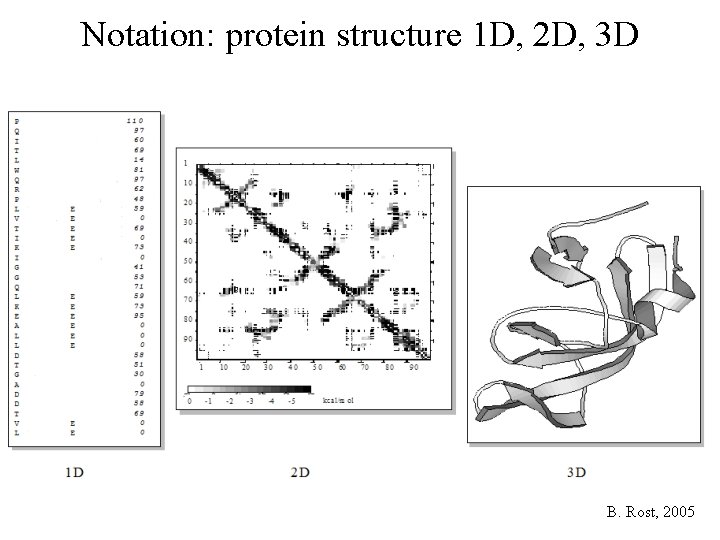
Notation: protein structure 1 D, 2 D, 3 D B. Rost, 2005

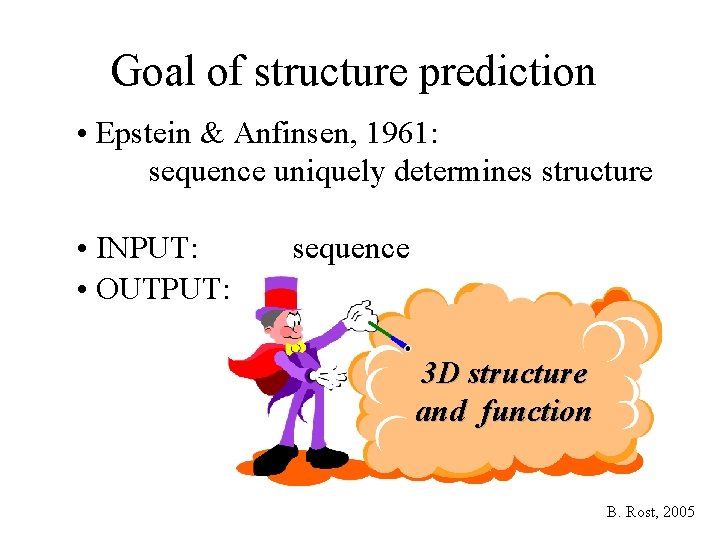
Goal of structure prediction • Epstein & Anfinsen, 1961: sequence uniquely determines structure • INPUT: • OUTPUT: sequence 3 D structure and function B. Rost, 2005

CASP – Olympics of Protein Structure Prediction • Critical Assessment of Techniques of Protein Structure Prediction • 1994, 1996, 1998, 2000, 20 02, 2004, 2006, 2008, 2010, 2012 • Blind Test, Independent Evaluation • CASP 9: 116 targets • CASP ROLL

1 D Structure Prediction • • Secondary structure Solvent accessibility Disordered regions Domain boundary

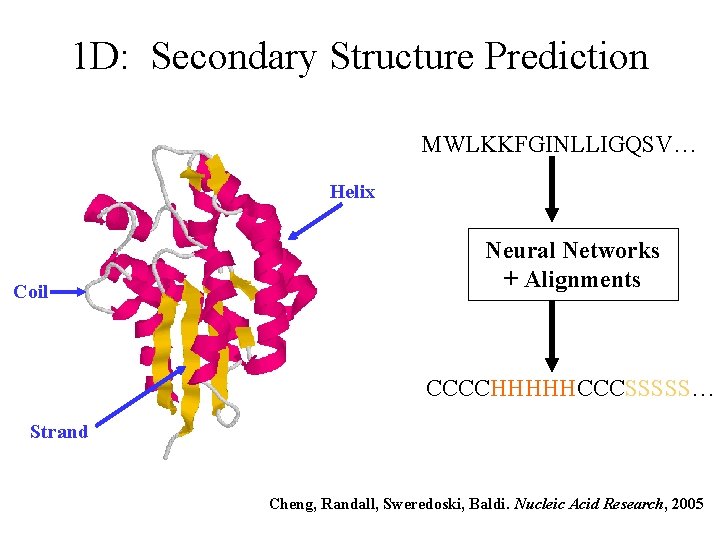
1 D: Secondary Structure Prediction MWLKKFGINLLIGQSV… Helix Coil Neural Networks + Alignments CCCCHHHHHCCCSSSSS… Strand Cheng, Randall, Sweredoski, Baldi. Nucleic Acid Research, 2005

Widely Used Tools (~78 -80%) SSpro 4. 1: http: //sysbio. rnet. missouri. edu/multicom_toolbox/ Distill: http: //distill. ucd. ie/porter/ PSI-PRED: http: //bioinf. cs. ucl. ac. uk/psipred/psiform. html software is also available SAM: http: //compbio. soe. ucsc. edu/SAM_T 08/T 08 -query. html PHD: http: //www. predictprotein. org/

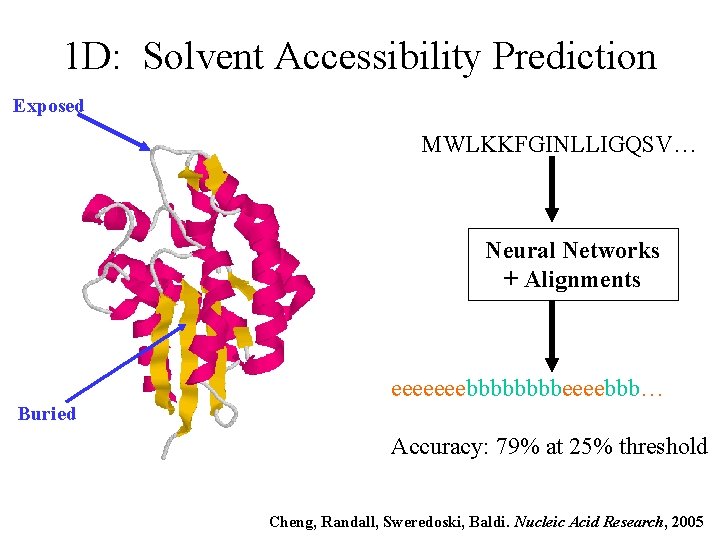
1 D: Solvent Accessibility Prediction Exposed MWLKKFGINLLIGQSV… Neural Networks + Alignments eeeeeeebbbbeeeebbb… Buried Accuracy: 79% at 25% threshold Cheng, Randall, Sweredoski, Baldi. Nucleic Acid Research, 2005

Widely Used Tools (78%) • ACCpro 4. 1: software: http: //sysbio. rnet. missouri. edu/multicom_toolbox/ • SCRATCH: http: //scratch. proteomics. uci. edu/ • PHD: http: //www. predictprotein. org/ • Distill: http: //distill. ucd. ie/porter/

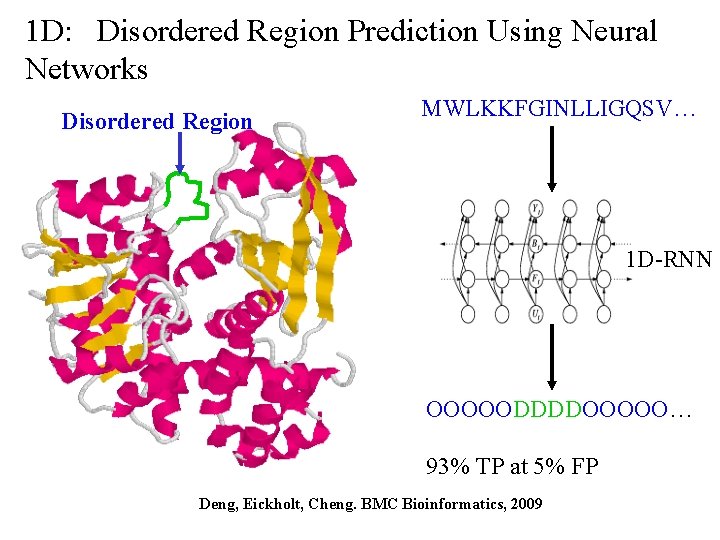
1 D: Disordered Region Prediction Using Neural Networks Disordered Region MWLKKFGINLLIGQSV… 1 D-RNN OOOOODDDDOOOOO… 93% TP at 5% FP Deng, Eickholt, Cheng. BMC Bioinformatics, 2009

Tools Pre. Disorder: http: //sysbio. rnet. missouri. edu/multicom_toolbox/ A collection of disorder predictors: http: //www. disprot. org/predictors. php Deng, Eickholt, Cheng. BMC Bioinformatics, 2009 & Mol. Biosystem, 2011

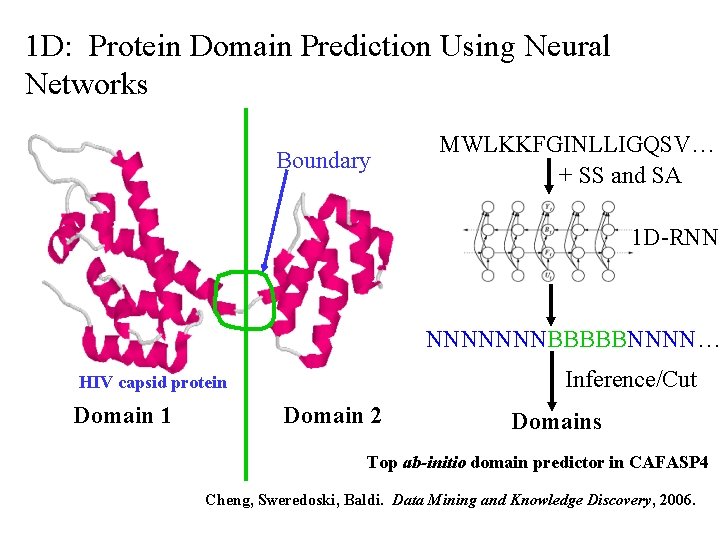
1 D: Protein Domain Prediction Using Neural Networks Boundary MWLKKFGINLLIGQSV… + SS and SA 1 D-RNN NNNNNNNBBBBBNNNN… Inference/Cut HIV capsid protein Domain 1 Domain 2 Domains Top ab-initio domain predictor in CAFASP 4 Cheng, Sweredoski, Baldi. Data Mining and Knowledge Discovery, 2006.

Web: http: //sysbio. rnet. missouri. edu/multicom_toolbox/index. html


Project 1 – CASP ROLL • Predict Secondary Structure, Solvent Accessibility, Disorder Regions of five CASP ROLL targets • Data: http: //predictioncenter. org /casprol/targetlist. cgi • Make predictions for R 001, R 002, R 003, R 005, R 006

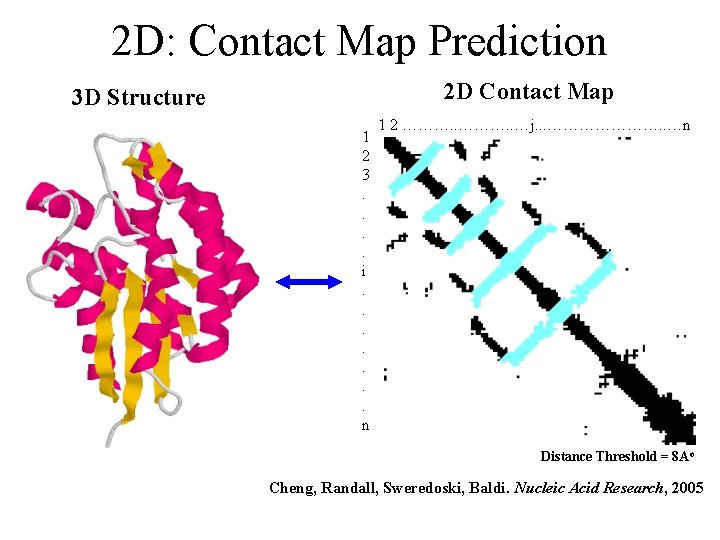
2 D: Contact Map Prediction 3 D Structure 2 D Contact Map 1 2 ………. . …j. . . …………………. . …n 1 2 3. . i. . . . n Distance Threshold = 8 Ao Cheng, Randall, Sweredoski, Baldi. Nucleic Acid Research, 2005

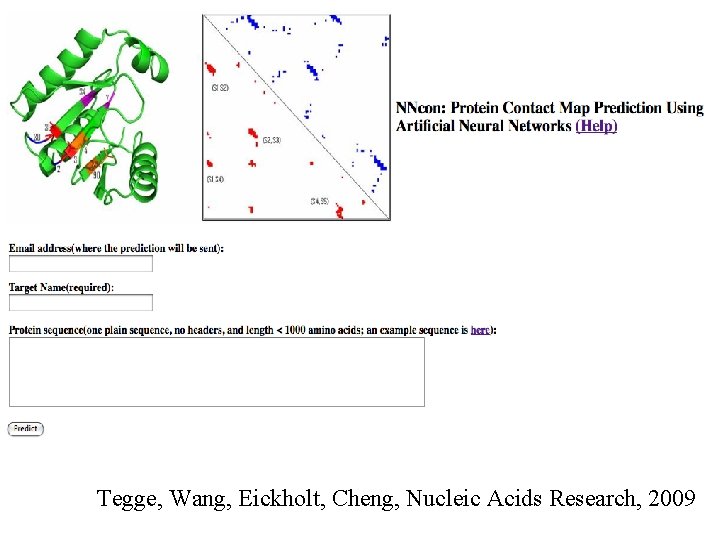
Contact Prediction • SVMcon: http: //casp. rnet. missouri. edu/svmcon. html • NNcon: http: //casp. rnet. missouri. edu/nncon. html • SCRATCH: http: //scratch. proteomics. uci. edu/ • SAM: http: //compbio. soe. ucsc. edu/HMMapps/HMM-applications. html

Tegge, Wang, Eickholt, Cheng, Nucleic Acids Research, 2009

Two Methodologies for 3 D Structure Prediction • AB Initio Method (physical-chemical principles / molecular dynamics, knowledge -based (e. g. contact-based)) • Template-Based Method (knowledge-based approaches)

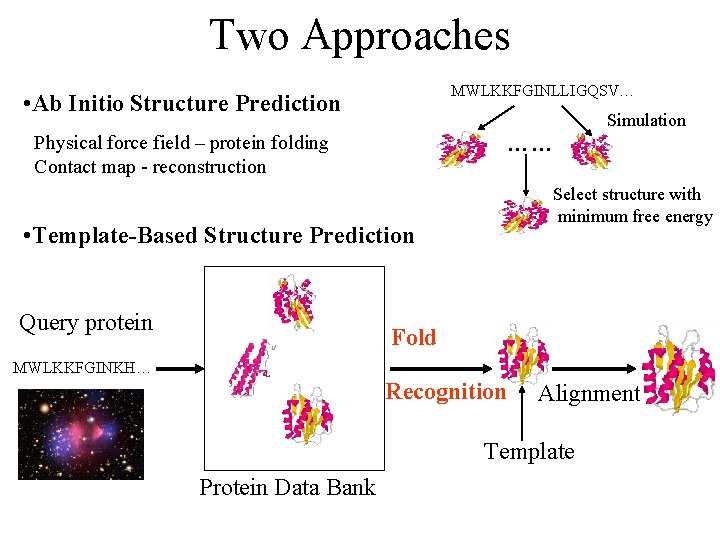
Two Approaches MWLKKFGINLLIGQSV… • Ab Initio Structure Prediction …… Physical force field – protein folding Contact map - reconstruction Select structure with minimum free energy • Template-Based Structure Prediction Query protein Simulation Fold MWLKKFGINKH… Recognition Alignment Template Protein Data Bank

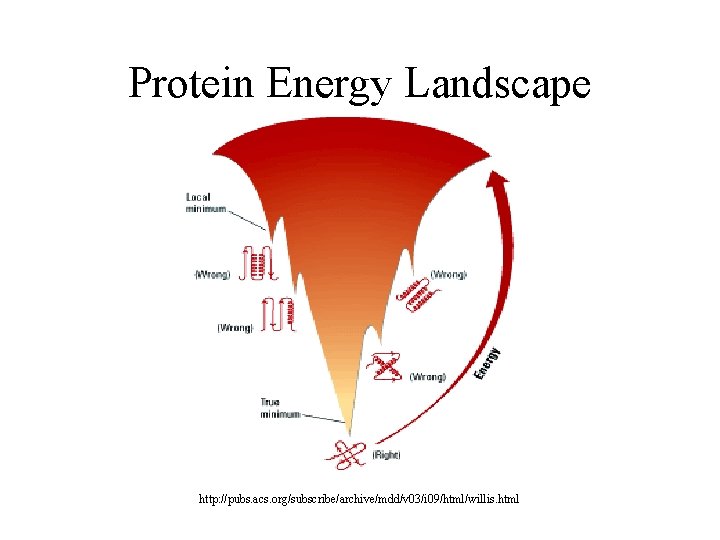
Protein Energy Landscape http: //pubs. acs. org/subscribe/archive/mdd/v 03/i 09/html/willis. html

Markov Chain Monte Carlo Simulation

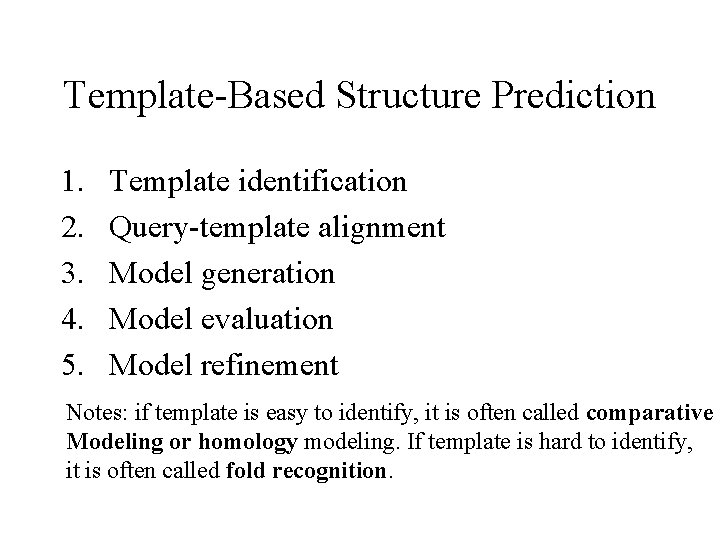
Template-Based Structure Prediction 1. 2. 3. 4. 5. Template identification Query-template alignment Model generation Model evaluation Model refinement Notes: if template is easy to identify, it is often called comparative Modeling or homology modeling. If template is hard to identify, it is often called fold recognition.

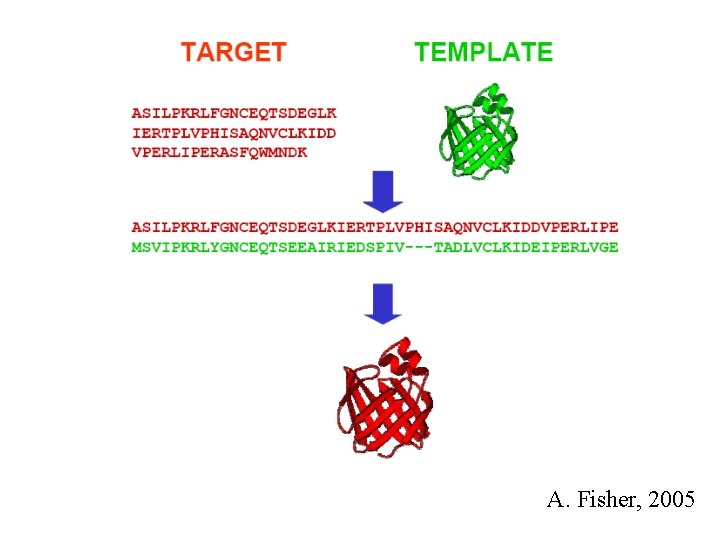
A. Fisher, 2005

Modeller • Need an alignment file between query and template sequence in the PIR format • Need the structure (atom coordinates) file of template protein • You need to write a simple script (Python for version 8. 2) to tell how to generate the model and where to find the alignment file and template structure file. • Run Modeller on the script. Modeller will automatically copy coordinates and make necessary adjustments to generate a model. • See project step 5 -8 for more details.

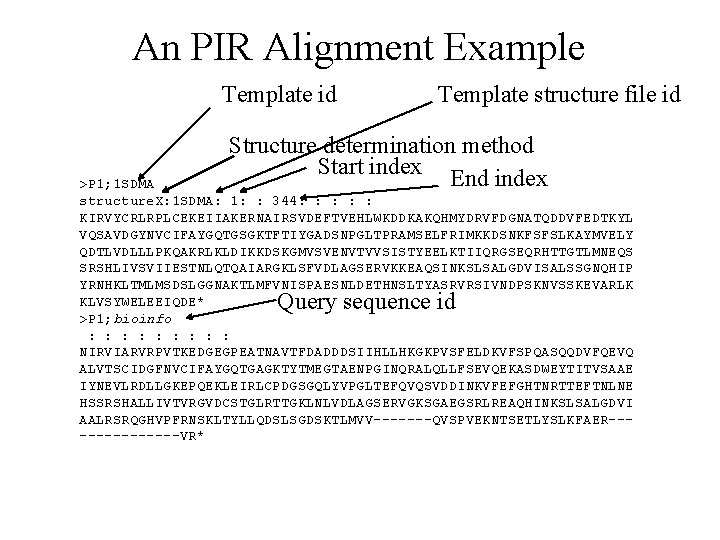
An PIR Alignment Example Template id Template structure file id Structure determination method Start index End index >P 1; 1 SDMA structure. X: 1 SDMA: 1: : 344: : : KIRVYCRLRPLCEKEIIAKERNAIRSVDEFTVEHLWKDDKAKQHMYDRVFDGNATQDDVFEDTKYL VQSAVDGYNVCIFAYGQTGSGKTFTIYGADSNPGLTPRAMSELFRIMKKDSNKFSFSLKAYMVELY QDTLVDLLLPKQAKRLKLDIKKDSKGMVSVENVTVVSISTYEELKTIIQRGSEQRHTTGTLMNEQS SRSHLIVSVIIESTNLQTQAIARGKLSFVDLAGSERVKKEAQSINKSLSALGDVISALSSGNQHIP YRNHKLTMLMSDSLGGNAKTLMFVNISPAESNLDETHNSLTYASRVRSIVNDPSKNVSSKEVARLK KLVSYWELEEIQDE* >P 1; bioinfo : : : : : NIRVIARVRPVTKEDGEGPEATNAVTFDADDDSIIHLLHKGKPVSFELDKVFSPQASQQDVFQEVQ ALVTSCIDGFNVCIFAYGQTGAGKTYTMEGTAENPGINQRALQLLFSEVQEKASDWEYTITVSAAE IYNEVLRDLLGKEPQEKLEIRLCPDGSGQLYVPGLTEFQVQSVDDINKVFEFGHTNRTTEFTNLNE HSSRSHALLIVTVRGVDCSTGLRTTGKLNLVDLAGSERVGKSGAEGSRLREAQHINKSLSALGDVI AALRSRQGHVPFRNSKLTYLLQDSLSGDSKTLMVV-------QVSPVEKNTSETLYSLKFAER-------VR* Query sequence id

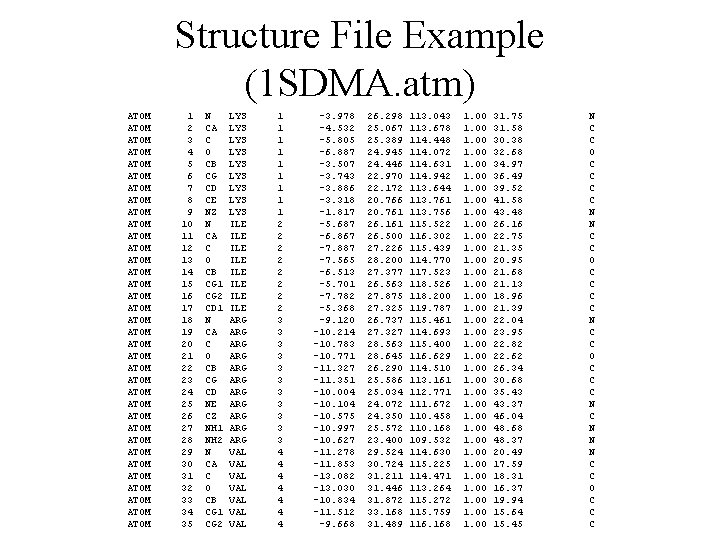
Structure File Example (1 SDMA. atm) ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 N CA C O CB CG CD CE NZ N CA C O CB CG 1 CG 2 CD 1 N CA C O CB CG CD NE CZ NH 1 NH 2 N CA C O CB CG 1 CG 2 LYS LYS LYS ILE ILE ARG ARG ARG VAL VAL 1 1 1 1 1 2 2 2 2 3 3 3 4 4 4 4 -3. 978 -4. 532 -5. 805 -6. 887 -3. 507 -3. 743 -3. 886 -3. 318 -1. 817 -5. 687 -6. 867 -7. 887 -7. 565 -6. 513 -5. 701 -7. 782 -5. 368 -9. 120 -10. 214 -10. 783 -10. 771 -11. 327 -11. 351 -10. 004 -10. 104 -10. 575 -10. 997 -10. 627 -11. 278 -11. 853 -13. 082 -13. 030 -10. 834 -11. 512 -9. 668 26. 298 25. 067 25. 389 24. 945 24. 446 22. 970 22. 172 20. 766 20. 761 26. 161 26. 500 27. 226 28. 200 27. 377 26. 563 27. 875 27. 325 26. 737 27. 327 28. 563 28. 645 26. 290 25. 586 25. 034 24. 072 24. 350 25. 572 23. 400 29. 524 30. 724 31. 211 31. 446 31. 872 33. 168 31. 489 113. 043 113. 678 114. 448 114. 072 114. 631 114. 942 113. 644 113. 761 113. 756 115. 522 116. 302 115. 439 114. 770 117. 523 118. 526 118. 200 119. 787 115. 461 114. 693 115. 400 116. 629 114. 510 113. 161 112. 771 111. 672 110. 458 110. 168 109. 532 114. 630 115. 225 114. 471 113. 264 115. 272 115. 759 116. 168 1. 00 1. 00 1. 00 31. 75 31. 58 30. 38 32. 68 34. 97 36. 49 39. 52 41. 58 43. 48 26. 16 22. 75 21. 35 20. 95 21. 68 21. 13 18. 96 21. 39 22. 04 23. 95 22. 82 22. 62 26. 34 30. 68 35. 43 43. 37 46. 04 48. 68 48. 37 20. 49 17. 59 18. 31 16. 37 19. 94 15. 64 15. 45 N C C O C C C C N C C O C C C N N N C C O C C C

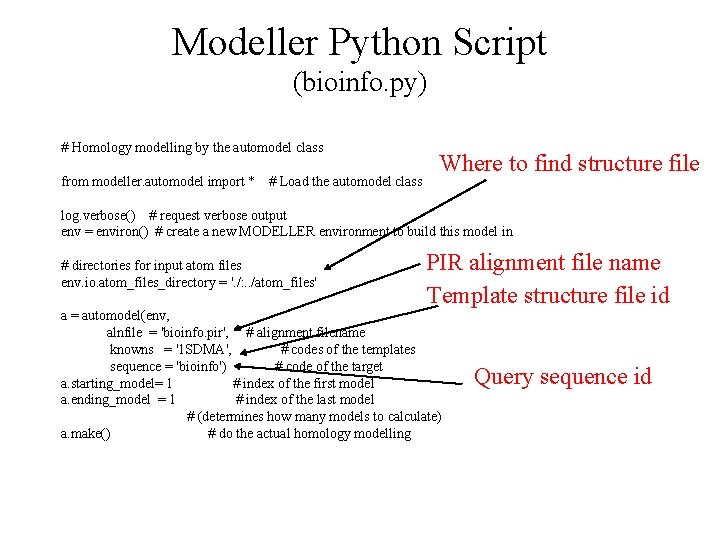
Modeller Python Script (bioinfo. py) # Homology modelling by the automodel class from modeller. automodel import * # Load the automodel class Where to find structure file log. verbose() # request verbose output env = environ() # create a new MODELLER environment to build this model in # directories for input atom files env. io. atom_files_directory = '. /: . . /atom_files' PIR alignment file name Template structure file id a = automodel(env, alnfile = 'bioinfo. pir', # alignment filename knowns = '1 SDMA', # codes of the templates sequence = 'bioinfo') # code of the target a. starting_model= 1 # index of the first model a. ending_model = 1 # index of the last model # (determines how many models to calculate) a. make() # do the actual homology modelling Query sequence id

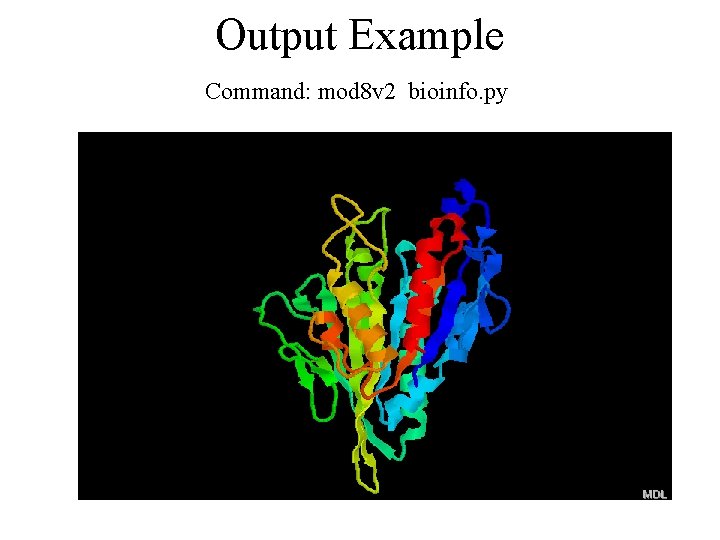
Output Example Command: mod 8 v 2 bioinfo. py

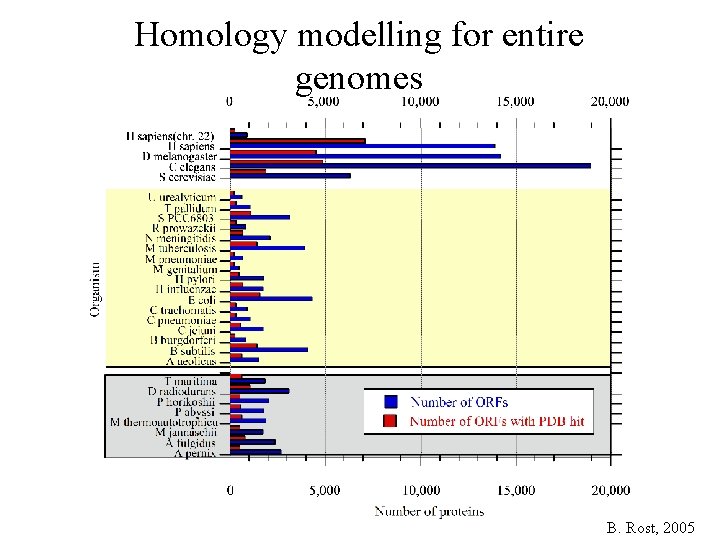
Homology modelling for entire genomes B. Rost, 2005

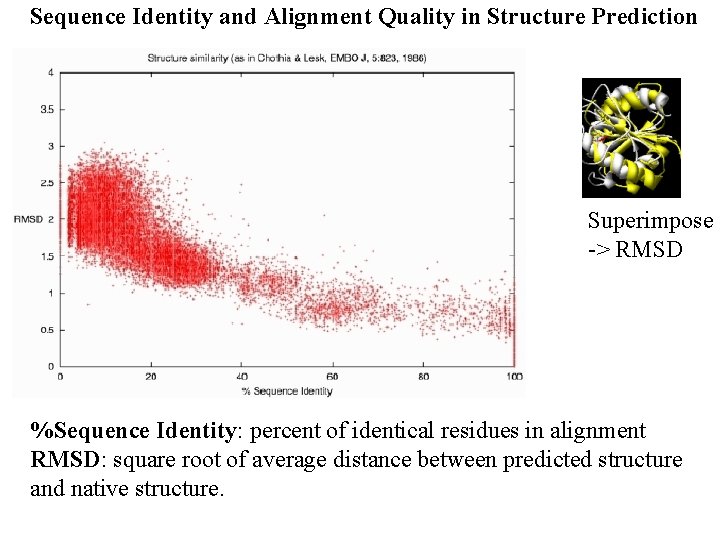
Sequence Identity and Alignment Quality in Structure Prediction Superimpose -> RMSD %Sequence Identity: percent of identical residues in alignment RMSD: square root of average distance between predicted structure and native structure.

3 D Structure Prediction Tools • MULTICOM (http: //sysbio. rnet. missouri. edu/multicom_toolbox/index. html ) • I-TASSER (http: //zhang. bioinformatics. ku. edu/I-TASSER/) • HHpred (http: //protevo. eb. tuebingen. mpg. de/toolkit/index. php? view=hhpred) • Robetta (http: //robetta. bakerlab. org/) • 3 D-Jury (http: //bioinfo. pl/Meta/) • FFAS (http: //ffas. ljcrf. edu/ffas-cgi/ffas. pl) • Pcons (http: //pcons. net/) • Sparks (http: //phyyz 4. med. buffalo. edu/hzhou/anonymous-foldsp 3. html) • FUGUE (http: //www-cryst. bioc. cam. ac. uk/%7 Efugue/prfsearch. html) • FOLDpro (http: //mine 5. ics. uci. edu: 1026/foldpro. html) • SAM (http: //www. cse. ucsc. edu/research/compbio/sam. html) • Phyre (http: //www. sbg. bio. ic. ac. uk/~phyre/) • 3 D-PSSM (http: //www. sbg. bio. ic. ac. uk/3 dpssm/) • m. Gen. Threader (http: //bioinf. cs. ucl. ac. uk/psipred/psiform. html)

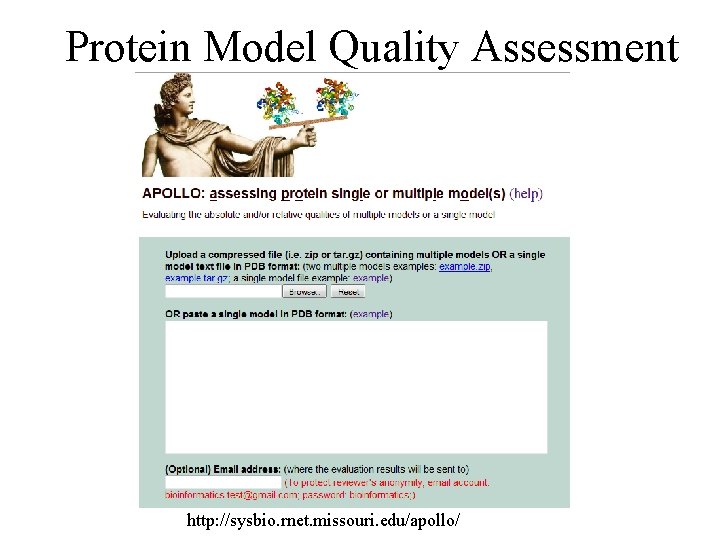
Protein Model Quality Assessment http: //sysbio. rnet. missouri. edu/apollo/

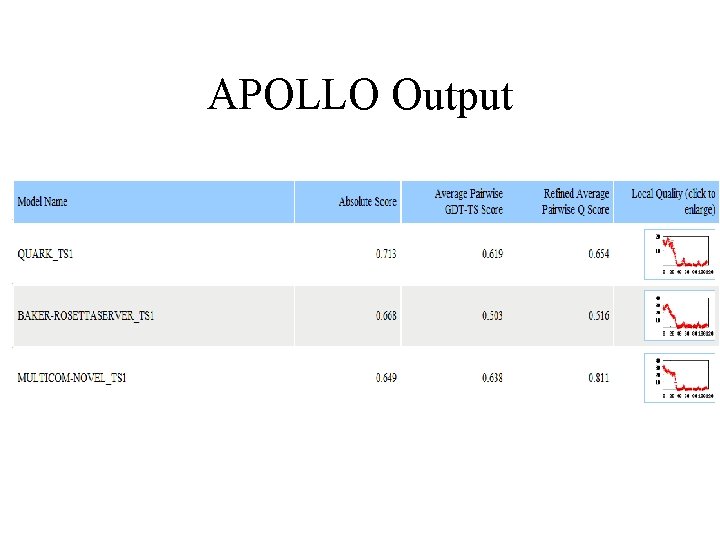
APOLLO Output

Application of Structure Prediction • Structure prediction is improving • Template-based structure become more and more practical. Particularly, comparative / homology modeling is pretty accurate in many cases. • Comparative modeling has been widely used in drug design. • Protein structure prediction (both secondary and tertiary) has become an indispensable tool of investigating function of proteins and mechanisms of biological processes.

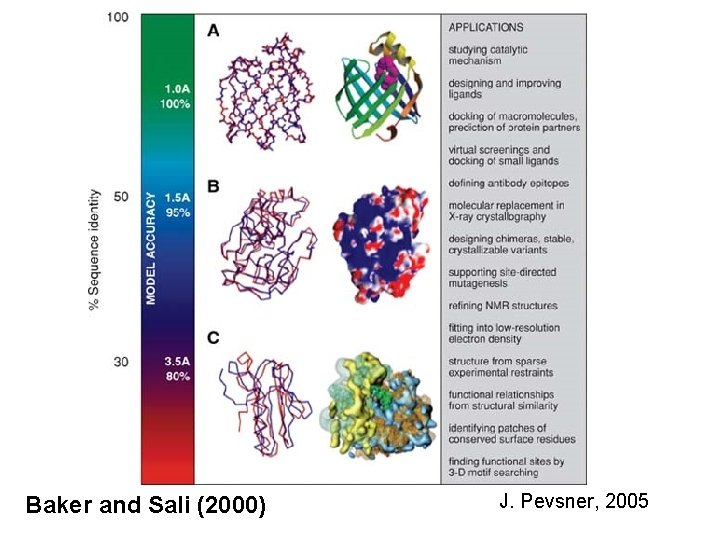
Baker and Sali (2000) J. Pevsner, 2005

Soy. KB: Genome-Wide Soybean Protein Structure Annotation Soybean genome Schmutz et al. , 2010 Joshi et al. , 2012

Project 2 – CASP 9 and CASP ROLL • Three CASP 9 targets (T 0584 -586) • Two CASP ROLL targets (R 001, R 002) • Predict 3 D structures • Visualize the structures CASP 9 targets: http: //predictioncenter. org/casp 9/targetlist. cgi CASP ROLL target list: http: //predictioncenter. org/casprol/targetlist. cgi