Protein Structure Determination by NMR Spectroscopy 1 D
Protein Structure Determination by NMR Spectroscopy
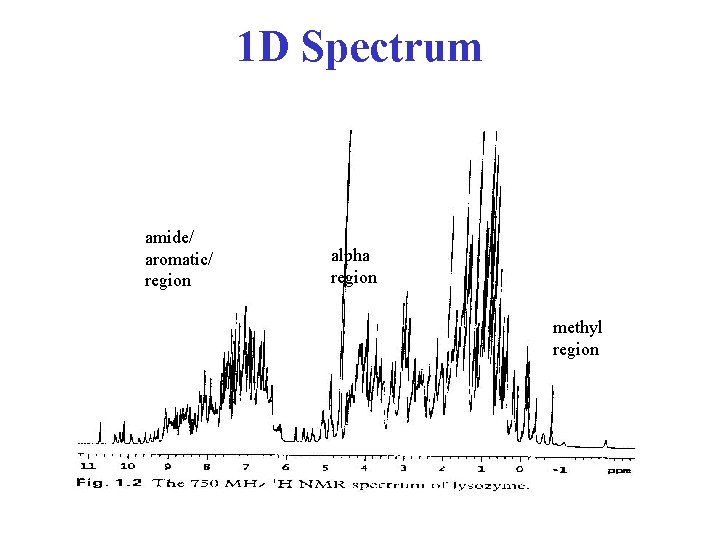
1 D Spectrum amide/ aromatic/ region alpha region methyl region
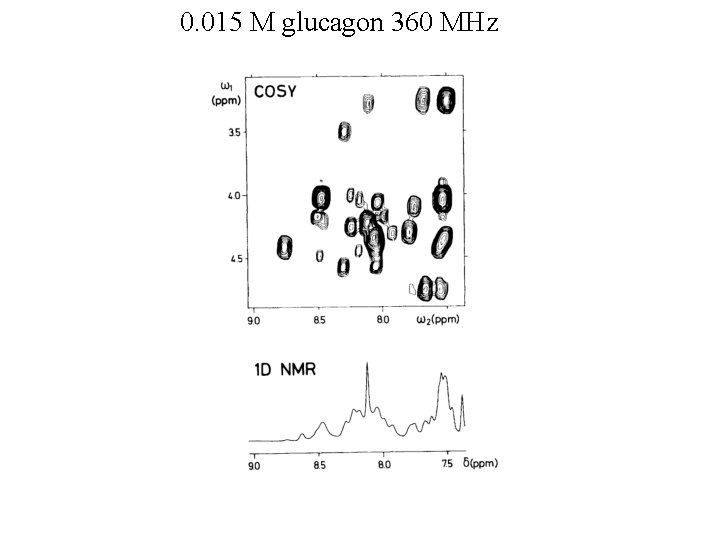
0. 015 M glucagon 360 MHz
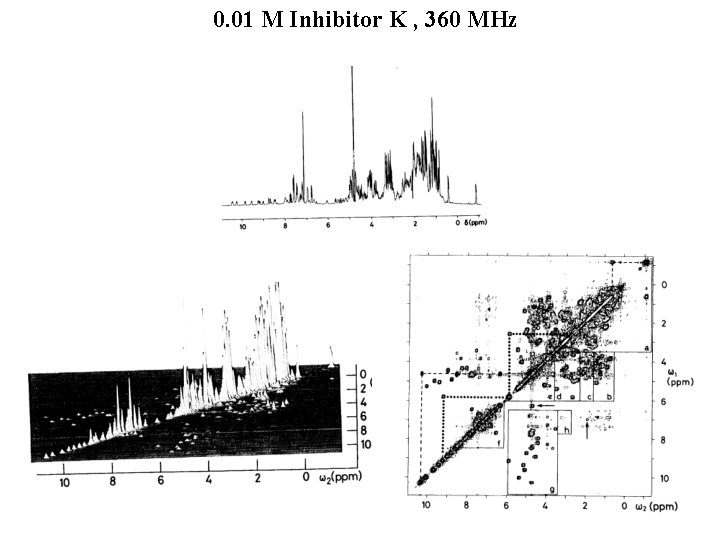
0. 01 M Inhibitor K , 360 MHz

2 D J-resolved COSY 45. COSY 90. COSY L. R. TOCSY Relayed COSY. (with one homo relayed). NOESY. ROESY.

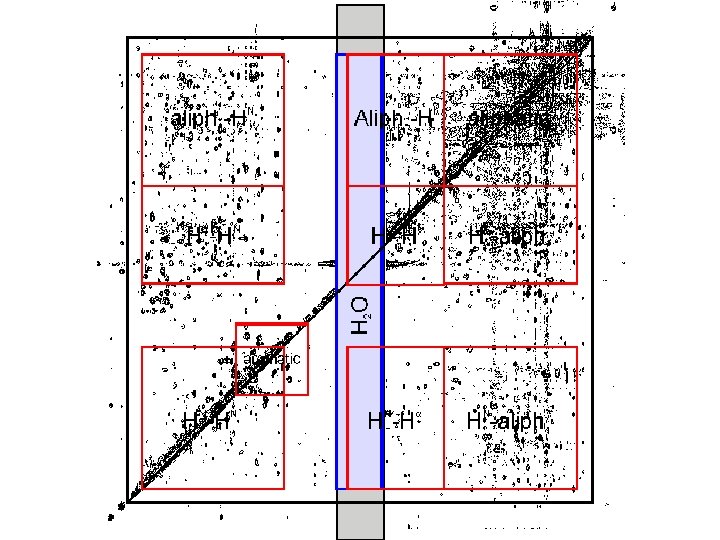
Protein Structure Determined by NMR 1 D NMR NOESY TOCSY DQF-COSY Peak Assignment Distance Dihedral angle Constraint Structural Simulation

Peak Assignment Strategies • Stage 1 Spin System Identification • Stage 2 Sequence-Specific Assignment
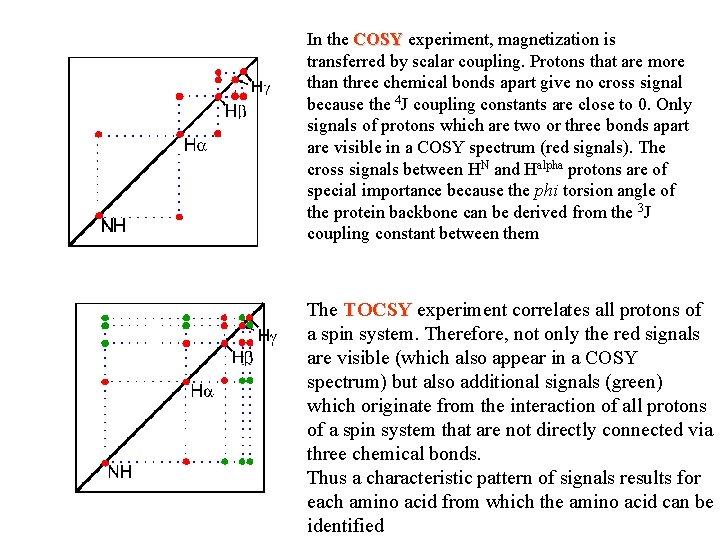
In the COSY experiment, magnetization is transferred by scalar coupling. Protons that are more than three chemical bonds apart give no cross signal because the 4 J coupling constants are close to 0. Only signals of protons which are two or three bonds apart are visible in a COSY spectrum (red signals). The cross signals between HN and Halpha protons are of special importance because the phi torsion angle of the protein backbone can be derived from the 3 J coupling constant between them The TOCSY experiment correlates all protons of a spin system. Therefore, not only the red signals are visible (which also appear in a COSY spectrum) but also additional signals (green) which originate from the interaction of all protons of a spin system that are not directly connected via three chemical bonds. Thus a characteristic pattern of signals results for each amino acid from which the amino acid can be identified
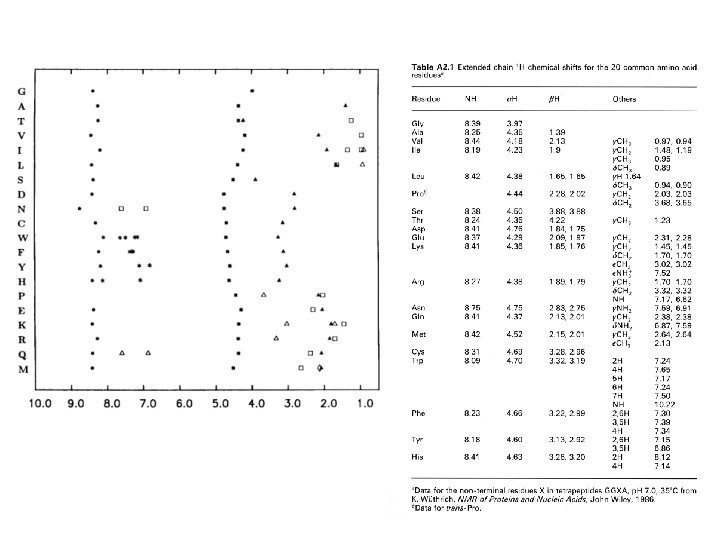

• approximate chemical shifts for various groups in a protein • “random coil” means “not having any specific structure” e. g. helix, sheet • measured in unstructured tetrapeptides • notice that none of these values is above 8. 8 (except Trp sidechain NH) or below 0. 9 • all the amides come between 8 and 8. 75 • alphas between 4 and 4. 8 • most methyls between 0. 9 and 1. 4 • some amino acids have identical spin systems and therefore identical signal patterns. They are: cysteine, aspartic acid, phenylalanine, histidine, asparagine, tryptophane and tyrosine ('AMX systems') on the one hand glutamic acid, glutamine and methionine ('AM(PT)X systems') on the other hand.
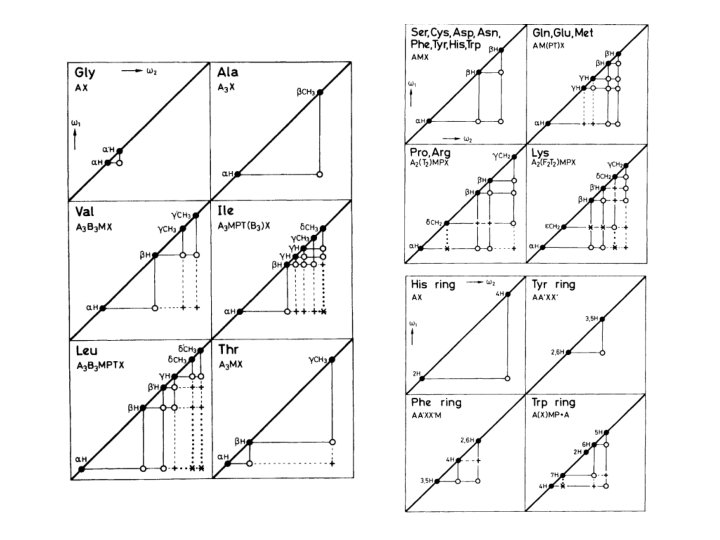
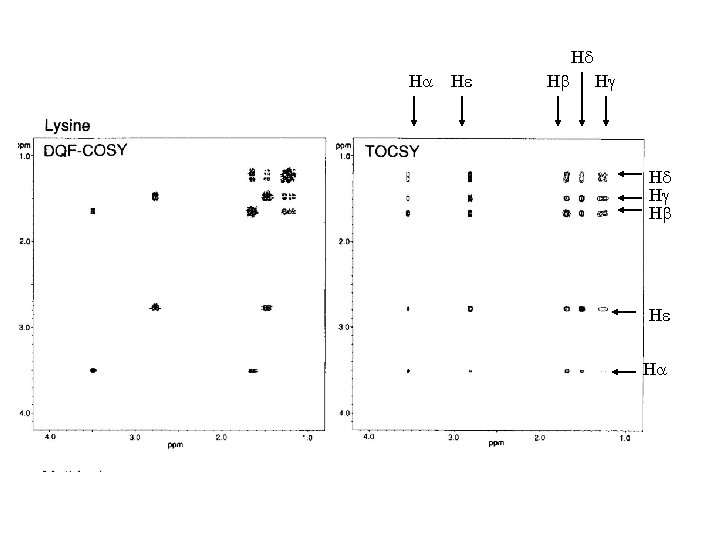
Hd Ha He Hb Hg Hd Hg Hb He Ha
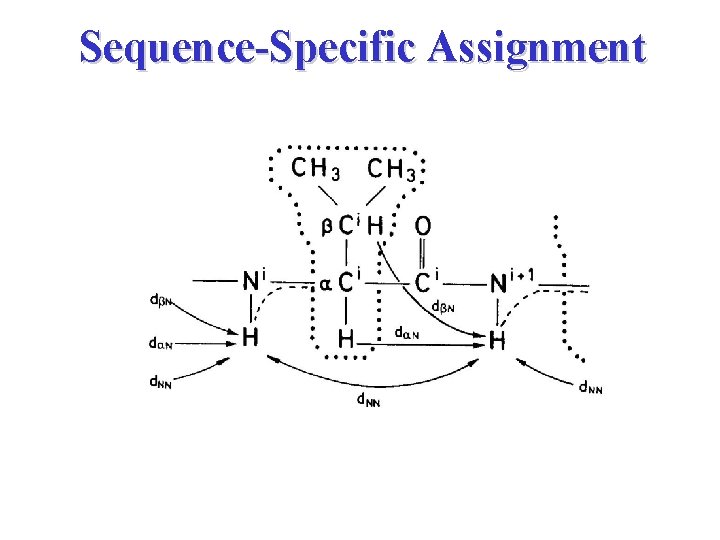
Sequence-Specific Assignment
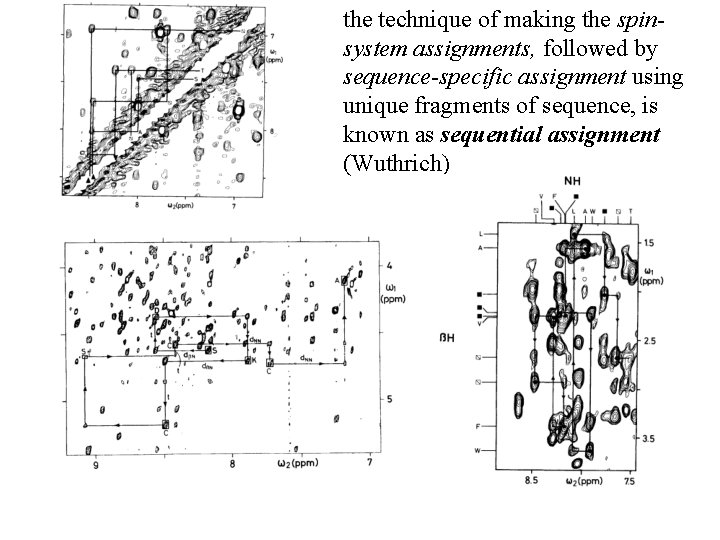
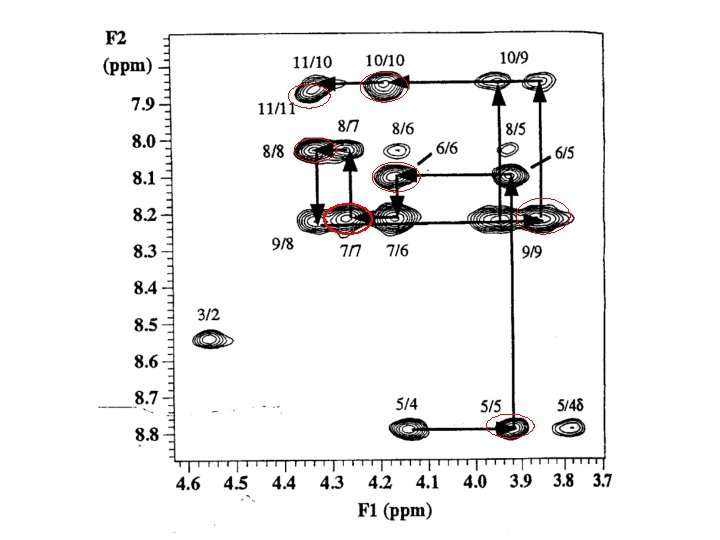
the technique of making the spinsystem assignments, followed by sequence-specific assignment using unique fragments of sequence, is known as sequential assignment (Wuthrich)

main-chain directed assignment (Englander). This technique does not focus on assigning all the spin systems first. Rather, it focuses on the backbone and links sizable stretches of backbone residues via sequential (i, i+1) n. Oe’s and other n. Oe’s that are characteristic of secondary structures. This technique is particularly useful when there is some knowledge of secondary structure beforehand.
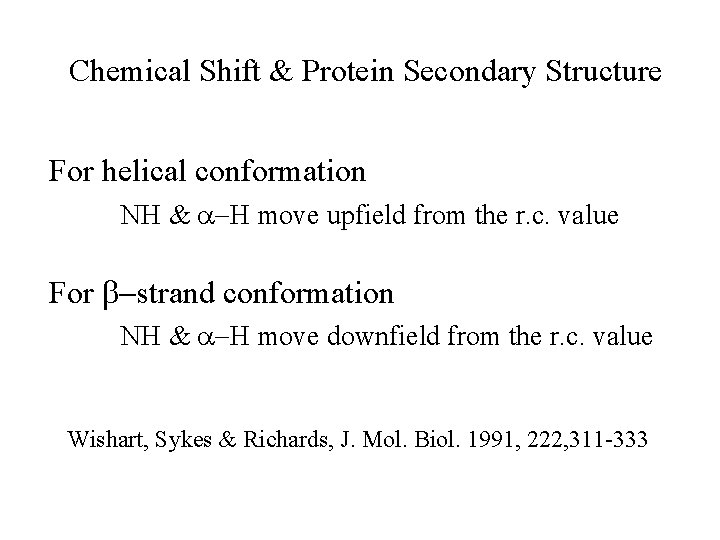
Chemical Shift & Protein Secondary Structure For helical conformation NH & a-H move upfield from the r. c. value For b-strand conformation NH & a-H move downfield from the r. c. value Wishart, Sykes & Richards, J. Mol. Biol. 1991, 222, 311 -333
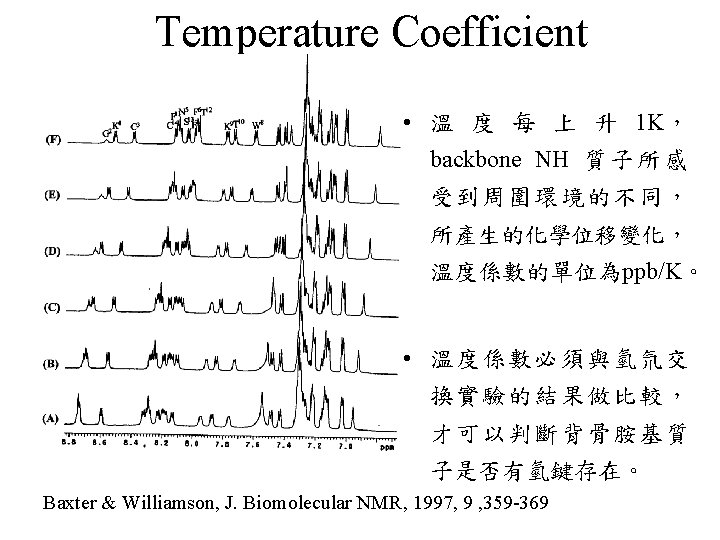
Backbone H-D Exchange 2 hour 30 min gp-41 fusion peptide in SDS micelle, 298 K
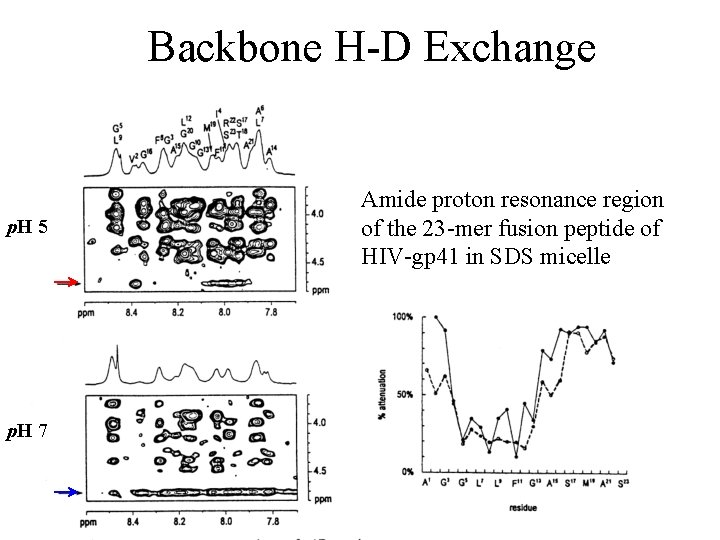
Backbone H-D Exchange p. H 5 p. H 7 Amide proton resonance region of the 23 -mer fusion peptide of HIV-gp 41 in SDS micelle
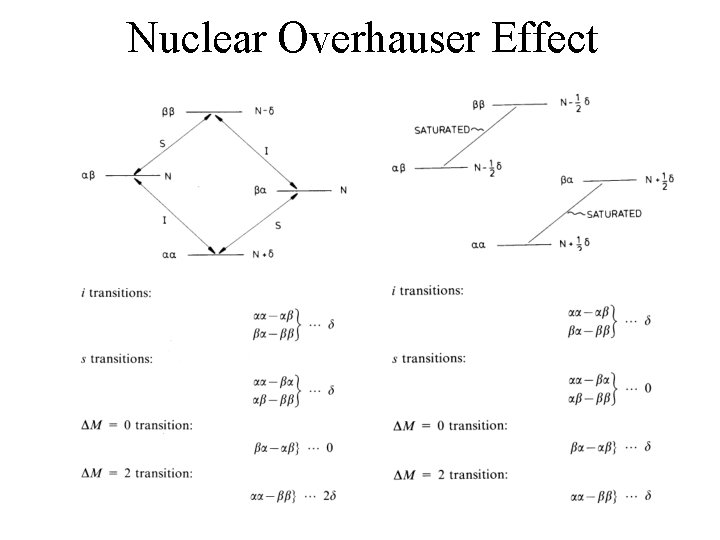
Nuclear Overhauser Effect
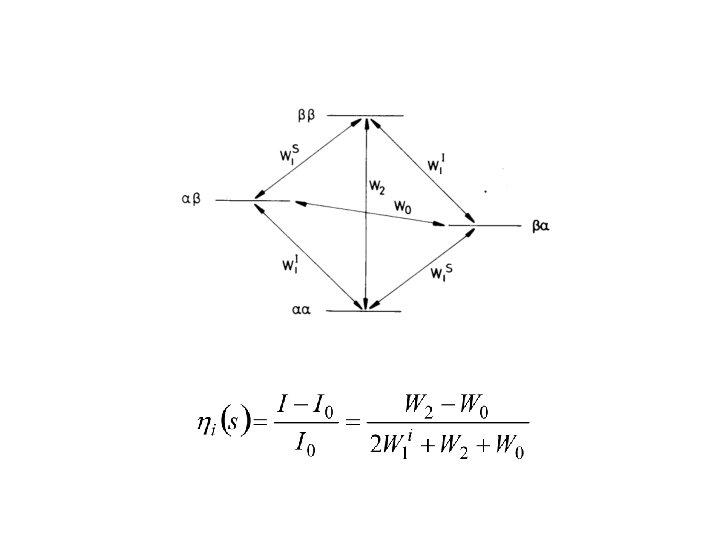
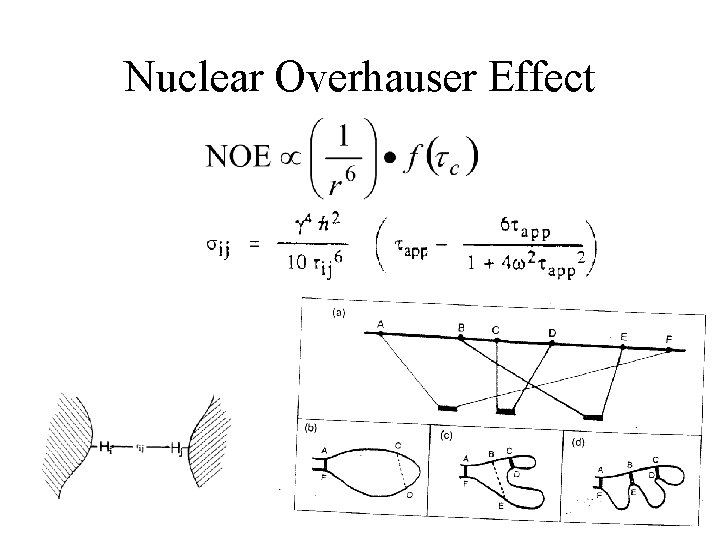
Nuclear Overhauser Effect
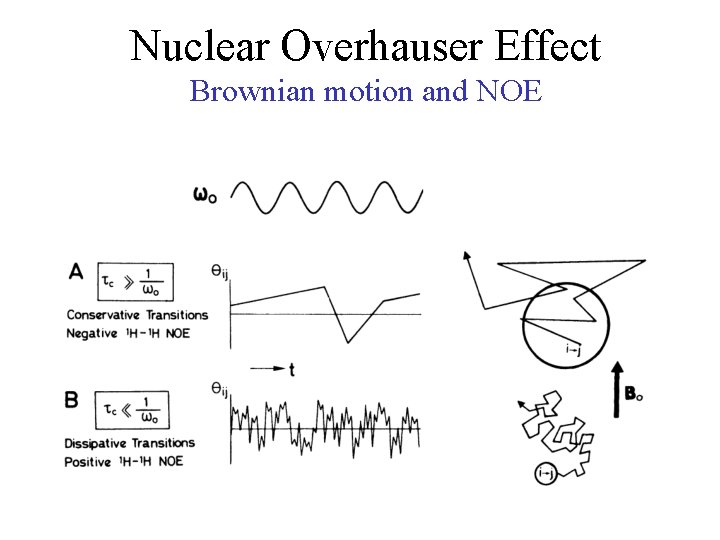
Nuclear Overhauser Effect Brownian motion and NOE
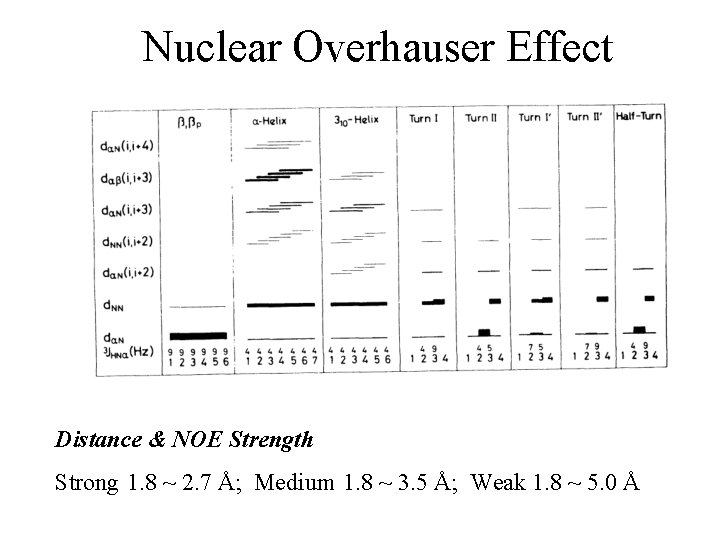
Nuclear Overhauser Effect Distance & NOE Strength Strong 1. 8 ~ 2. 7 Å; Medium 1. 8 ~ 3. 5 Å; Weak 1. 8 ~ 5. 0 Å
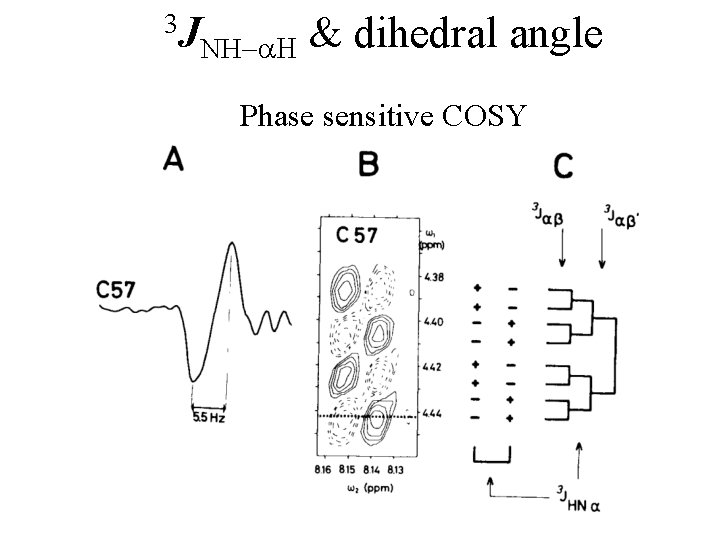
3 J NH-a. H & dihedral angle Phase sensitive COSY
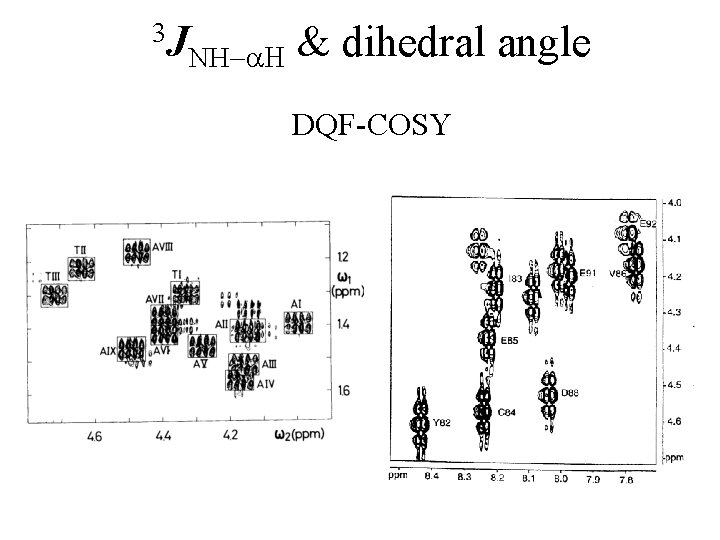
3 J NH-a. H & dihedral angle DQF-COSY
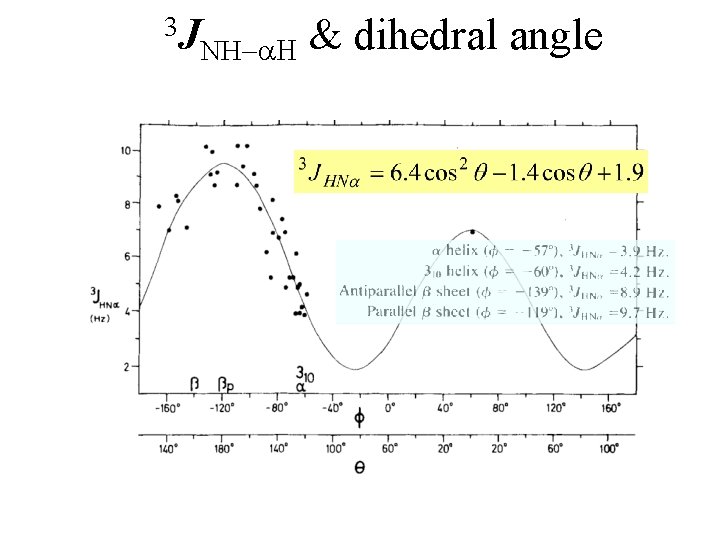
3 J NH-a. H & dihedral angle
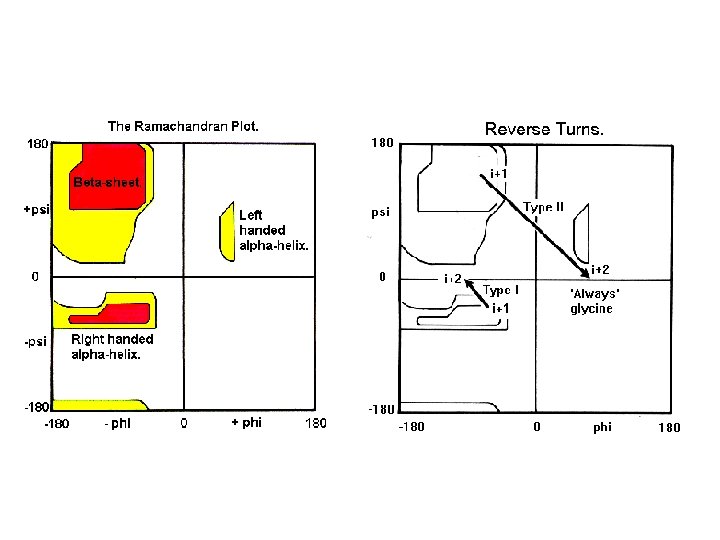
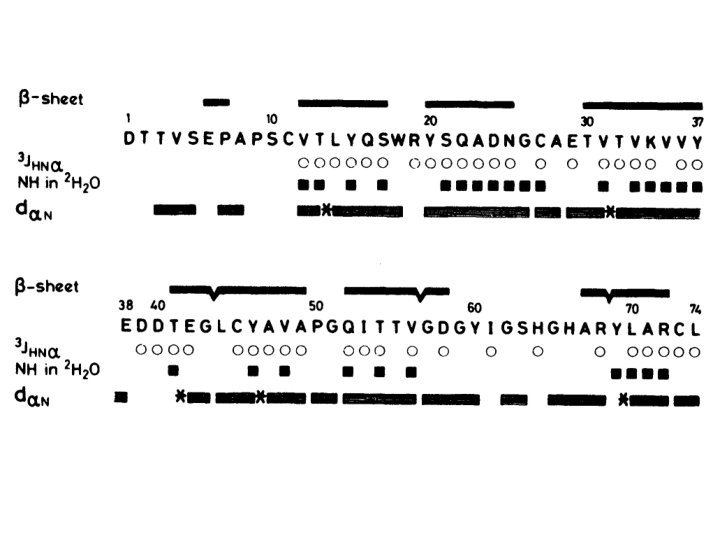
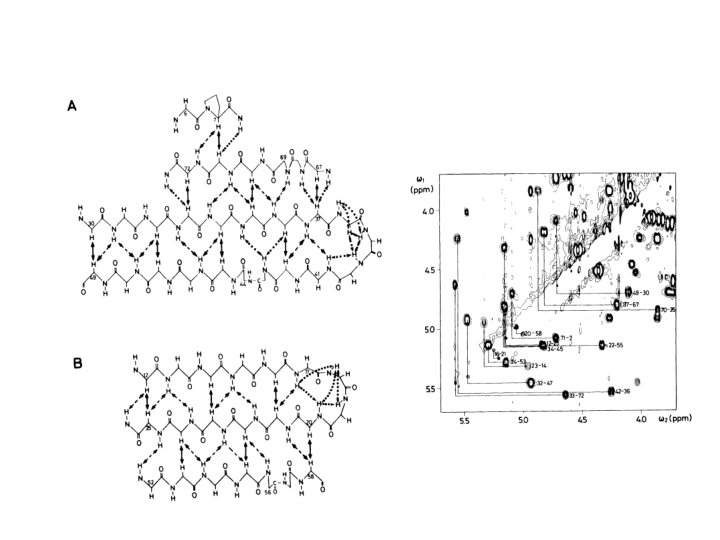
- Slides: 33