Protein structure databases visualization and comparison Protein structure

Protein structure databases, visualization, and comparison

Protein structure databases
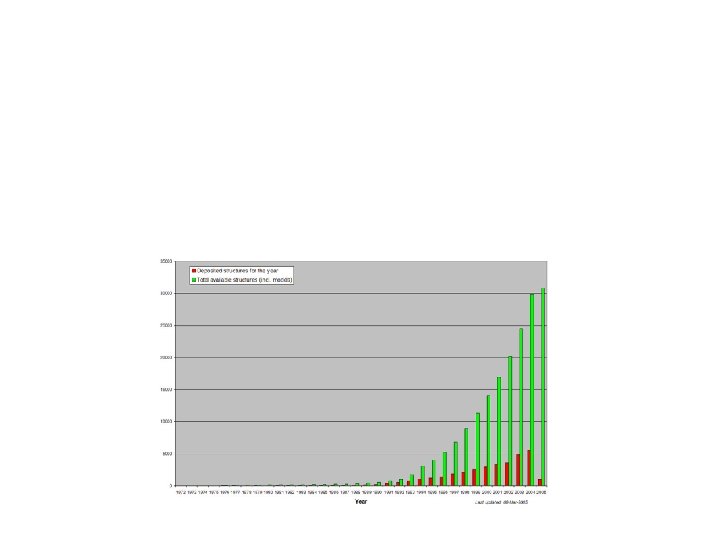



PDB entry

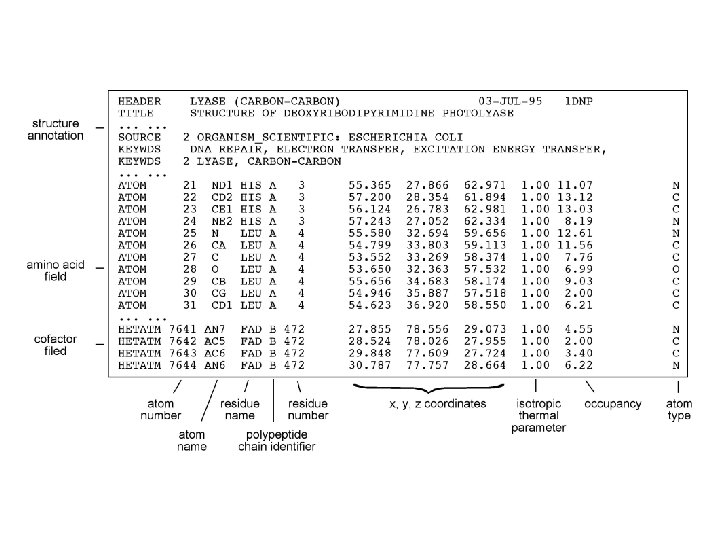
PDB format




mm. CIF and MMDB (optional)

mm. CIF



NCBI MMDB (optional)


Protein structure visualization



Molecular structure representation forms



Swiss-PDBViewer
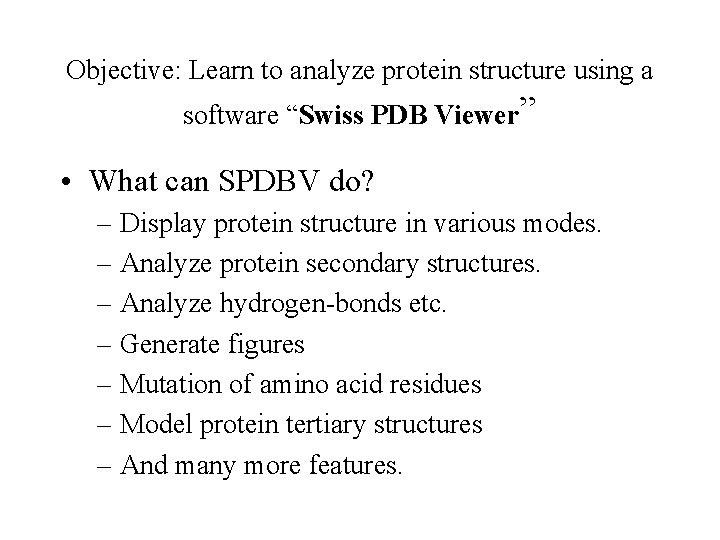
Objective: Learn to analyze protein structure using a software “Swiss PDB Viewer” • What can SPDBV do? – Display protein structure in various modes. – Analyze protein secondary structures. – Analyze hydrogen-bonds etc. – Generate figures – Mutation of amino acid residues – Model protein tertiary structures – And many more features.

Why SPDBV? • It is free, powerful, and user-friendly. • It runs on Windows and Apple computers. • It comes with on-line tutorials and manuals.

SPDBV usages • On line user manual: http: //expasy. org/spdbv/text/main. htm
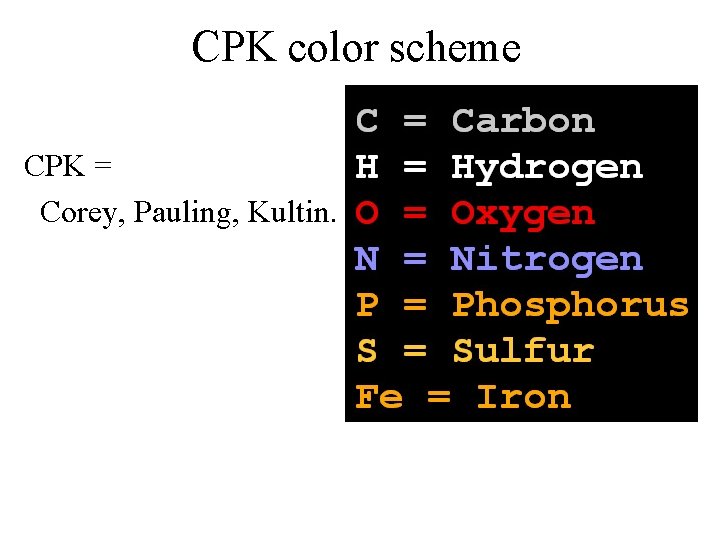
CPK color scheme CPK = Corey, Pauling, Kultin.
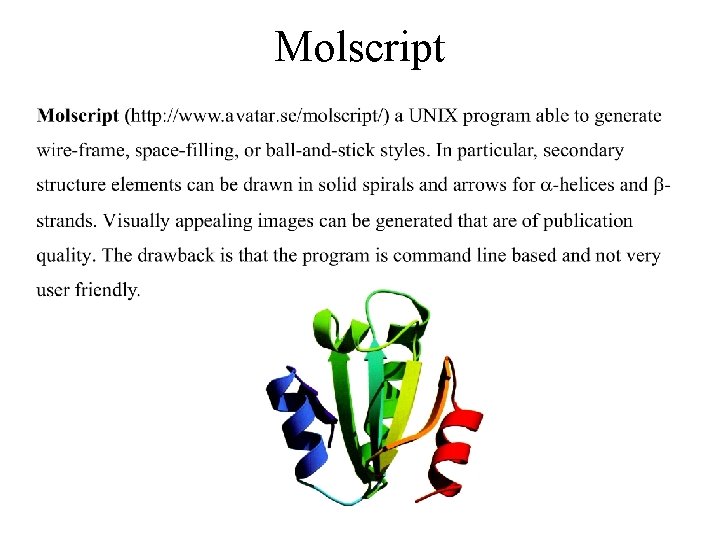
Molscript
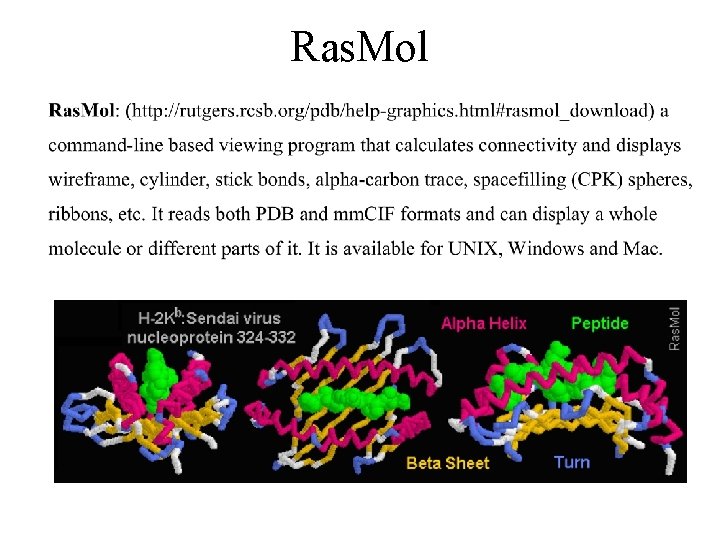
Ras. Mol
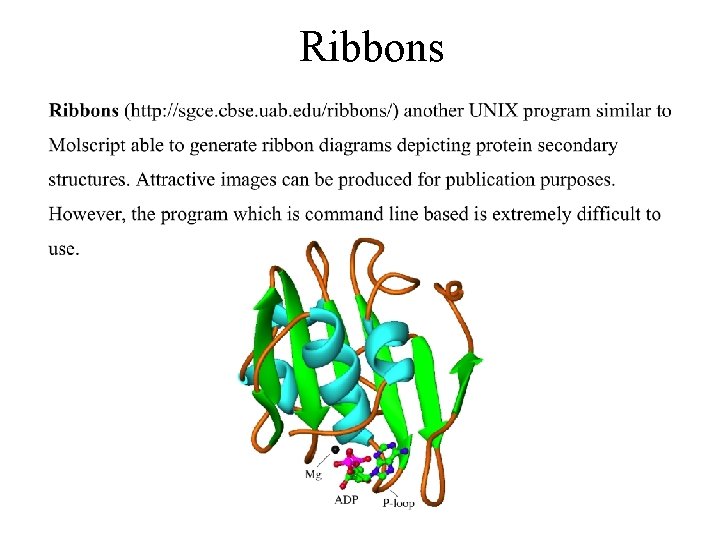
Ribbons
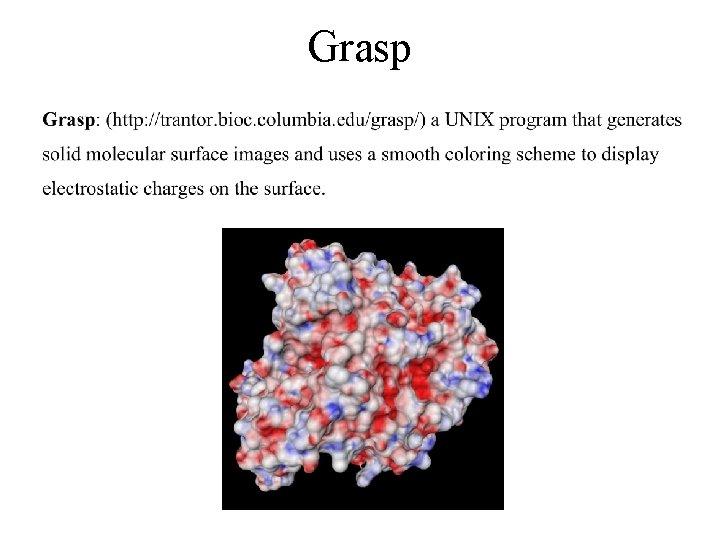
Grasp

Web-based visualization tools
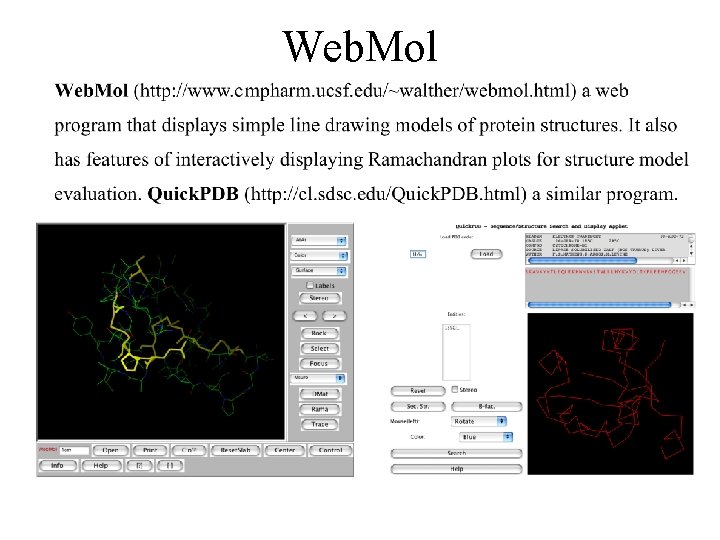
Web. Mol
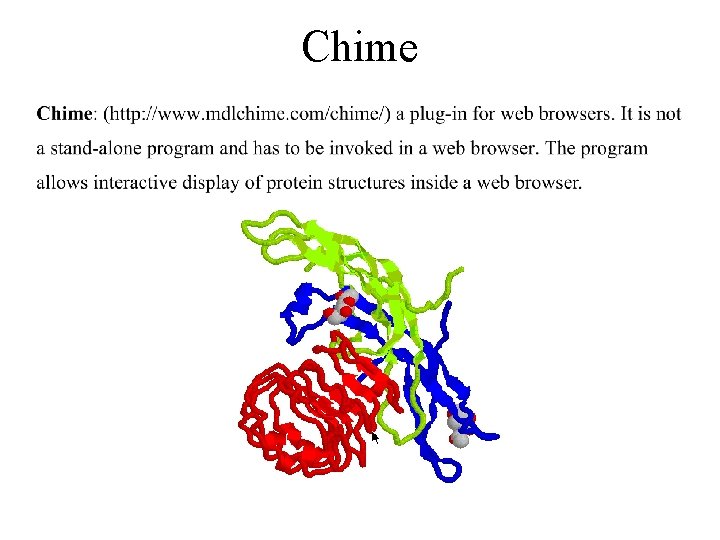
Chime
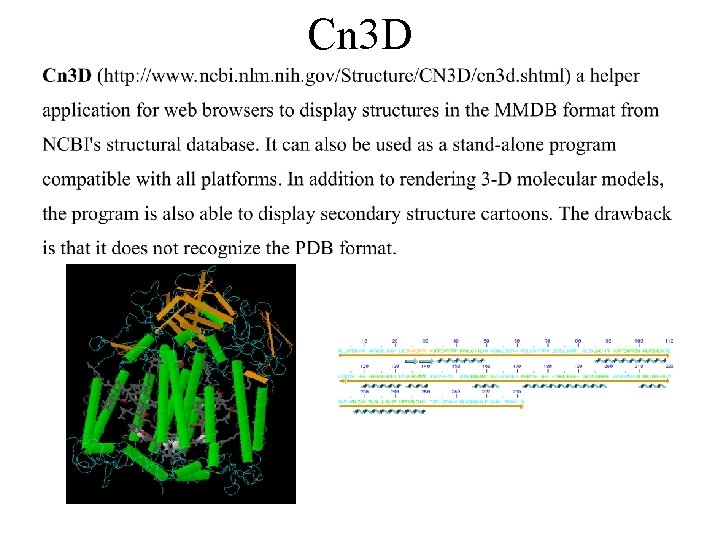
Cn 3 D
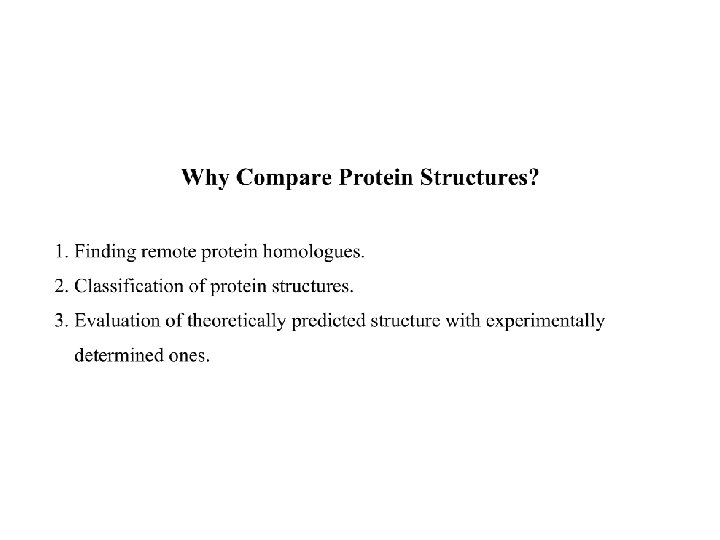
Protein structure comparison
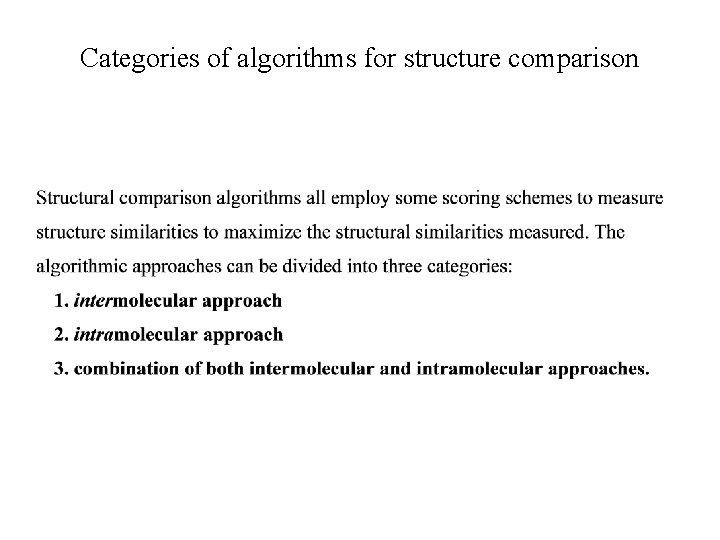
Categories of algorithms for structure comparison
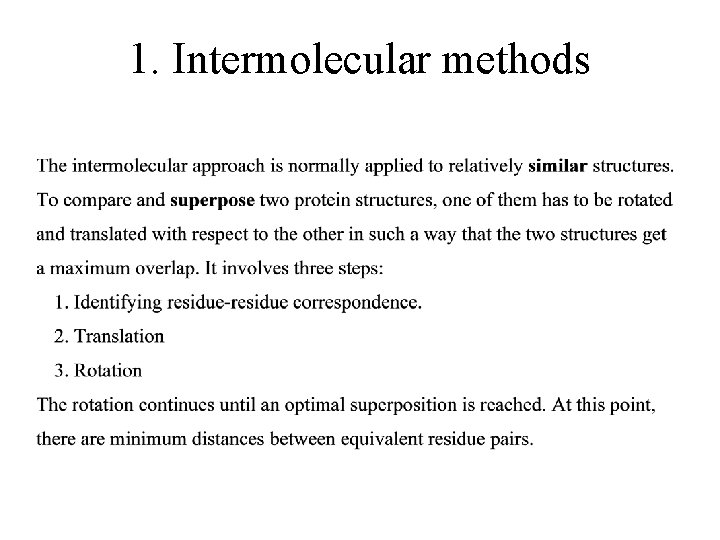
1. Intermolecular methods

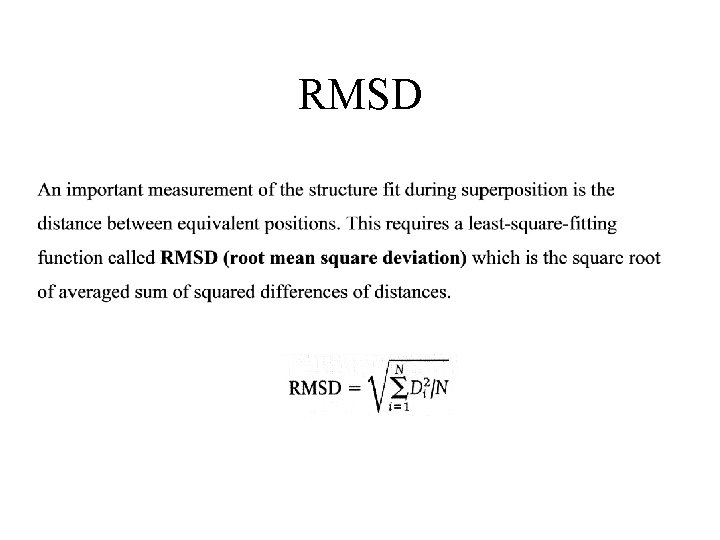
RMSD
2. Intramolecular methods to compare protein structures

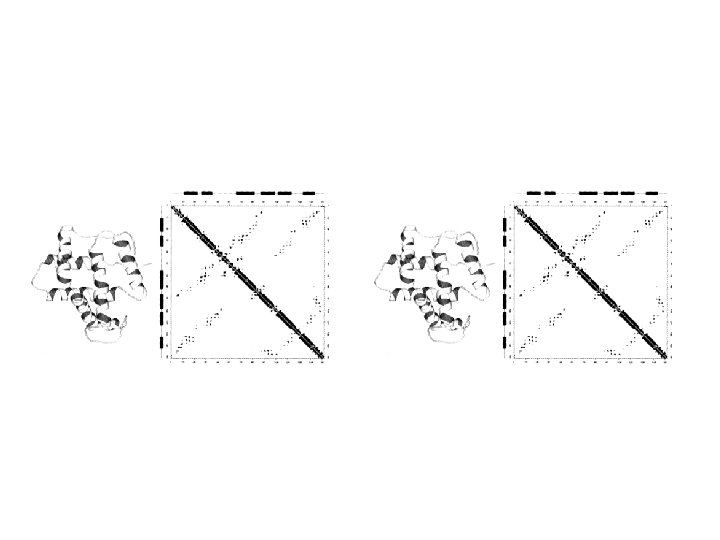

3. Combined methods
Multiple structure alignment

Protein Structure Classification
Steps of protein structure classification

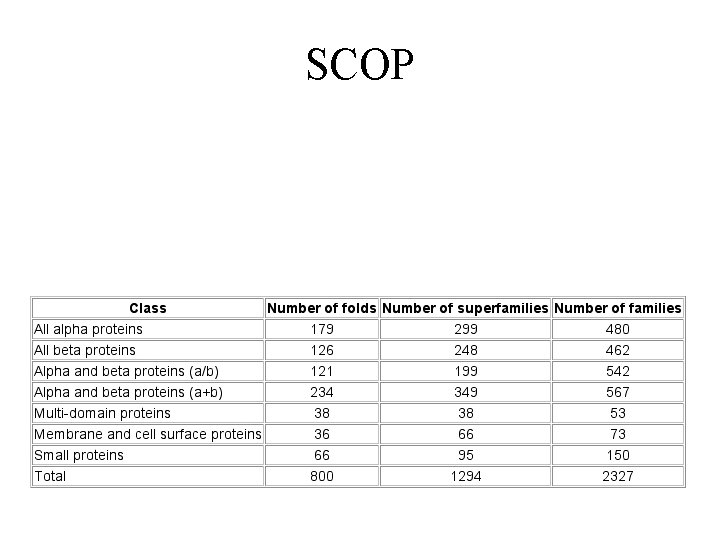
SCOP

SCOP definitions
Summary • PDB format • Protein structures are compared by 3 methods. – Inter-molecular • RMSD • Superimpose of molecules. – Intra-molecular (distance matrix) • SCOP is a manual curated database.

Reading assignments • Deep. View manual and online tutorials

- Slides: 55