Protein Structure Databases n Databases of three dimensional

Protein Structure Databases n Databases of three dimensional structures of proteins, where structure has been solved using X-ray crystallography or nuclear magnetic resonance (NMR) techniques n Protein Databases: n n PDB (protein data bank) Swiss-Prot PIR (Protein Information Resource) SCOP (Structural Classification of Proteins)

Protein Structure Databases n Most extensive for 3 -D structure is PDB
Visualization of Proteins n A number of programs convert atomic coordinates of 3 -d structures into views of the molecule n allow the user to manipulate the molecule by rotation, zooming, etc. n Critical in drug design -- yields insight into how the protein might interact with ligands at active sites

Visualization of Proteins Most popular programs for viewing 3 -D structures: Protein explorer: http: //www. umass. edu/microbio/chime/pe/protexpl/frntdoor. htm Rasmol: http: //www. umass. edu/microbio/rasmol/ Chime: http: //www. umass. edu/microbio/chime/ Cn 3 D: http: //www. ncbi. nlm. nih. gov/Structure/ Mage: http: //kinemage. biochem. duke. edu/website/kinhome. html Swiss 3 D viewer: http: //www. expasy. ch/spdbv/mainpage. html

Alignment of Protein Structure n Compare 3 D structure of one protein against 3 D structure of second protein n Compare positions of atoms in three-dimensional structures n Look for positions of secondary structural elements (helices and strands) within a protein domain n Exam distances between carbon atoms to determine degree structures may be superimposed n Side chain information can be incorporated n n Buried; visible Structural similarity between proteins does not necessarily mean evolutionary relationship

Alignment of Protein Structure
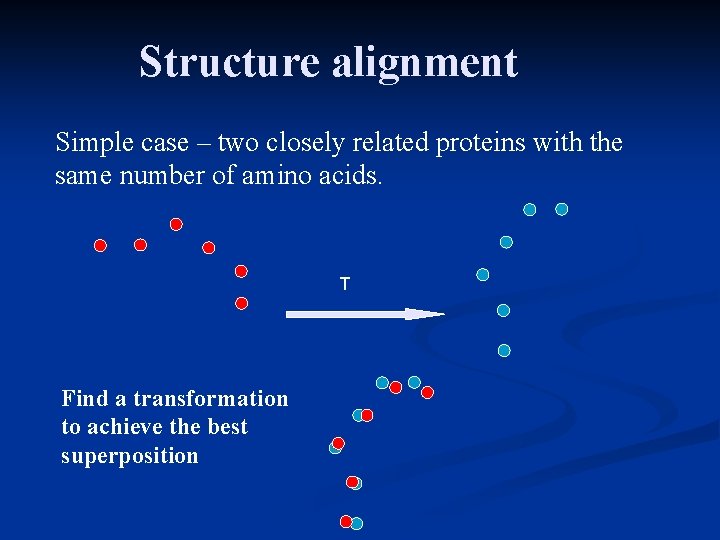
Structure alignment Simple case – two closely related proteins with the same number of amino acids. T Find a transformation to achieve the best superposition
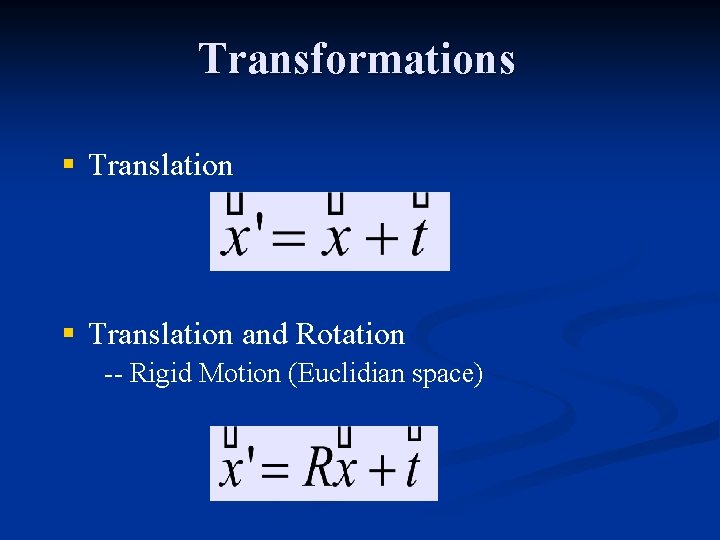
Transformations § Translation and Rotation -- Rigid Motion (Euclidian space)

Types of Structure Comparison § Sequence-dependent vs. sequence-independent structural alignment § Global vs. local structural alignment § Pairwise vs. multiple structural alignment

Sequence-dependent Structure Comparison 1234567 ASCRKLE ¦¦¦¦¦¦¦ ASCRKLE 2 1 3 4 6 5 7 2 1 4 5 3 7 6 Minimize rmsd of distances 1 -1, . . . , 7 -7 2 2 11 33 4 4 5 5 6 6 7 7

Sequence-dependent Structure Comparison n Can be solved in O(n) time. n Useful in comparing structures of the same protein solved in different methods, under different conformation, through dynamics. n Evaluation protein structure prediction.
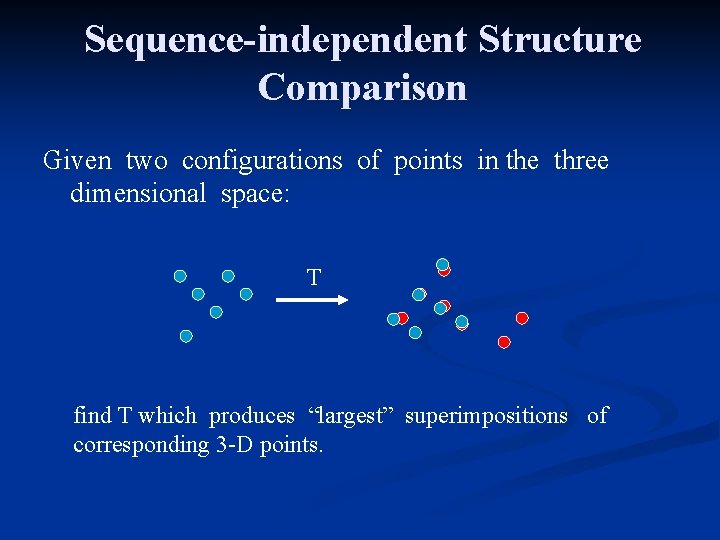
Sequence-independent Structure Comparison Given two configurations of points in the three dimensional space: T find T which produces “largest” superimpositions of corresponding 3 -D points.

Evaluating Structural Alignments 1. Number of amino acid correspondences created. 2. RMSD of corresponding amino acids 3. Percent identity in aligned residues 4. Number of gaps introduced 5. Size of the two proteins 6. Conservation of known active site environments 7. … No universally agreed upon criteria. It depends on what you are using the alignment for.

Protein Secondary Structure Prediction
Why secondary structure prediction? n n n Accurate secondary structure prediction can be an important information for the tertiary structure prediction Protein function prediction Protein classification Predicting structural change An easier problem than 3 D structure prediction (more than 40 years of history).
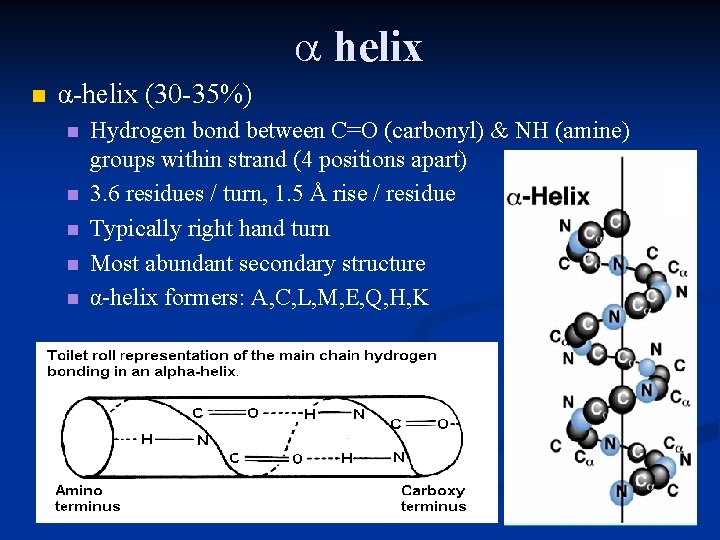
helix n α-helix (30 -35%) n n n Hydrogen bond between C=O (carbonyl) & NH (amine) groups within strand (4 positions apart) 3. 6 residues / turn, 1. 5 Å rise / residue Typically right hand turn Most abundant secondary structure α-helix formers: A, C, L, M, E, Q, H, K
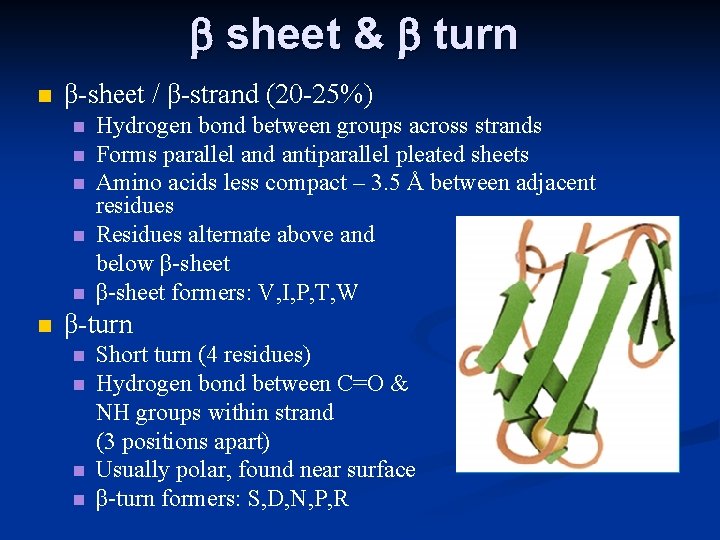
b sheet & b turn n β-sheet / β-strand (20 -25%) n n n Hydrogen bond between groups across strands Forms parallel and antiparallel pleated sheets Amino acids less compact – 3. 5 Å between adjacent residues Residues alternate above and below β-sheet formers: V, I, P, T, W β-turn n n Short turn (4 residues) Hydrogen bond between C=O & NH groups within strand (3 positions apart) Usually polar, found near surface β-turn formers: S, D, N, P, R

Others n Loop Regions between α-helices and β-sheets n On the surface, vary in length and 3 D configurations n Do not have regular periodic structures n Loop formers: small polar residues n n Coil (40 -50%) n Generally speaking, anything besides α-helix, βsheet, β-turn
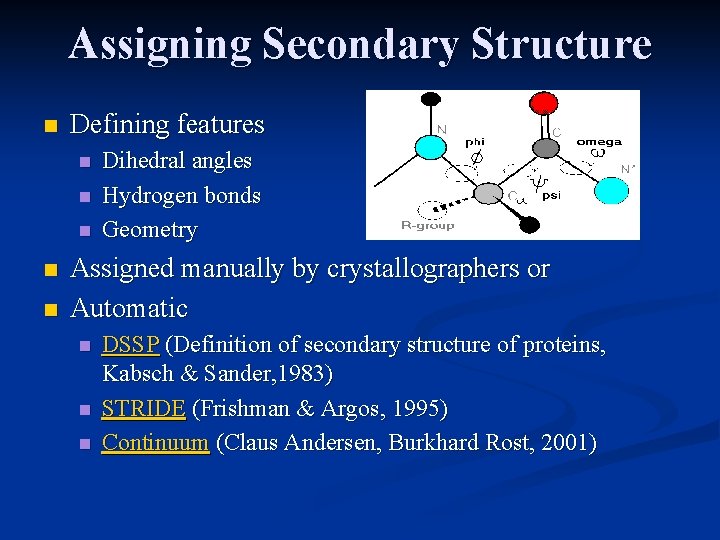
Assigning Secondary Structure n Defining features n n n Dihedral angles Hydrogen bonds Geometry Assigned manually by crystallographers or Automatic n n n DSSP (Definition of secondary structure of proteins, Kabsch & Sander, 1983) STRIDE (Frishman & Argos, 1995) Continuum (Claus Andersen, Burkhard Rost, 2001)

Definition of secondary structure of proteins (DSSP) n The DSSP code H = alpha helix n B = residue in isolated beta-bridge n E = extended strand, participates in beta ladder n G = 3 -helix (3/10 helix) n I = 5 helix (pi helix) n T = hydrogen bonded turn n S = bend n n CASP Standard n H = (H, G, I), E = (E, B), C = (T, S)

Secondary Structure Prediction n Given a protein sequence (primary structure) GHWIATR SSECPFIP HWIAT GQLIREAYEDYRHF GQLIREAYEDY SS n Predict its secondary structure content n (C=Coils H=Alpha Helix E=Beta Strands) GHWIATR SSECPFIP HWIAT GQLIREAYEDYRHF GQLIREAYEDY SS CEEEEEC HHCCCCCC EEEEE HHHHHHCCC HHHHHH HH

Algorithm n Chou-Fasman Method n Examining windows of 5 - 6 residues to predict structure
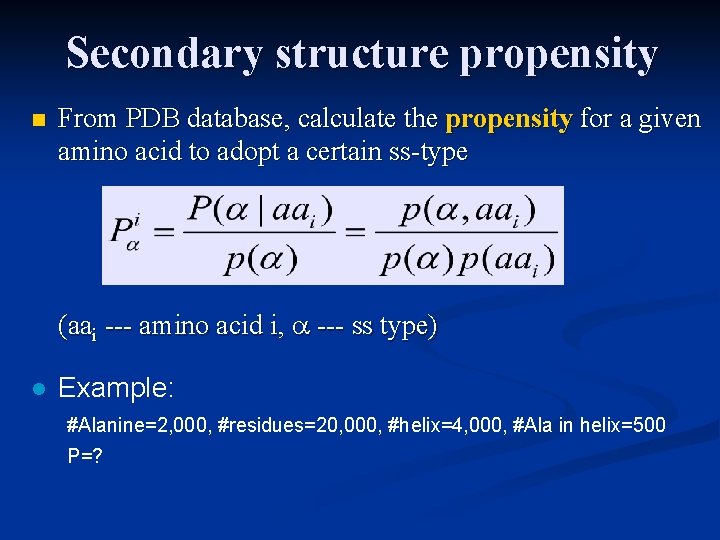
Secondary structure propensity n From PDB database, calculate the propensity for a given amino acid to adopt a certain ss-type (aai --- amino acid i, --- ss type) l Example: #Alanine=2, 000, #residues=20, 000, #helix=4, 000, #Ala in helix=500 P=?
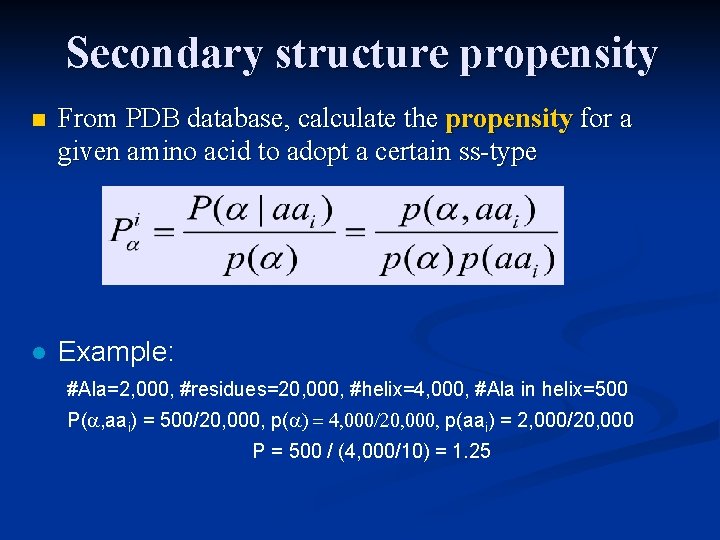
Secondary structure propensity n From PDB database, calculate the propensity for a given amino acid to adopt a certain ss-type l Example: #Ala=2, 000, #residues=20, 000, #helix=4, 000, #Ala in helix=500 P( , aai) = 500/20, 000, p( ) = 4, 000/20, 000, p(aai) = 2, 000/20, 000 P = 500 / (4, 000/10) = 1. 25

Chou-Fasman parameters Note: The parameters given in the textbook are 100*P i

Chou-Fasman algorithm n Helix: n n Scan through the peptide and identify regions where 4 out of 6 contiguous residues have P(H) > 1. 00. That region is declared an alpha-helix. Extend the helix in both directions until a set of four contiguous residues that have an average P(H) < 1. 00 is reached. That is declared the end of the helix. If the segment defined by this procedure is longer than 5 residues and the average P(H) > P(E) for that segment, the segment can be assigned as a helix. Repeat this procedure to locate all of the helical regions in the sequence.

Initiation Identify regions where 4/6 have a P(H) >1. 00 “alpha-helix nucleus” P(H) T S P T A E L M R S T G 69 77 57 69 142 151 121 145 98 77 69 57
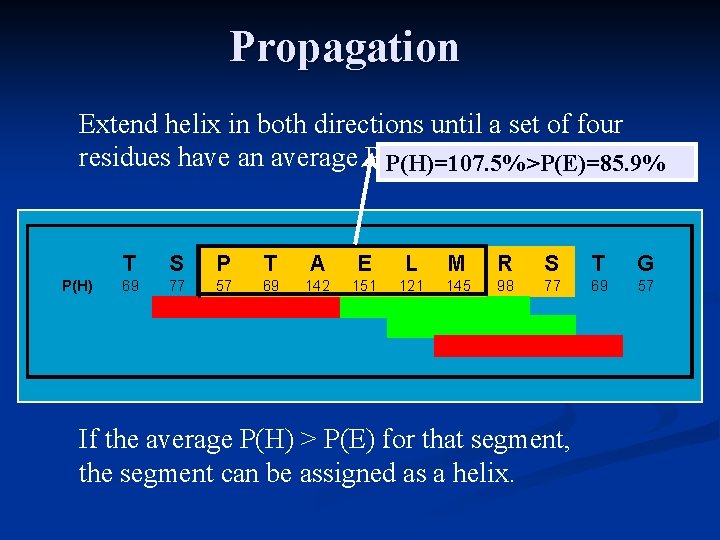
Propagation Extend helix in both directions until a set of four residues have an average P(H) <1. 00. P(H)=107. 5%>P(E)=85. 9% P(H) T S P T A E L M R S T G 69 77 57 69 142 151 121 145 98 77 69 57 If the average P(H) > P(E) for that segment, the segment can be assigned as a helix.
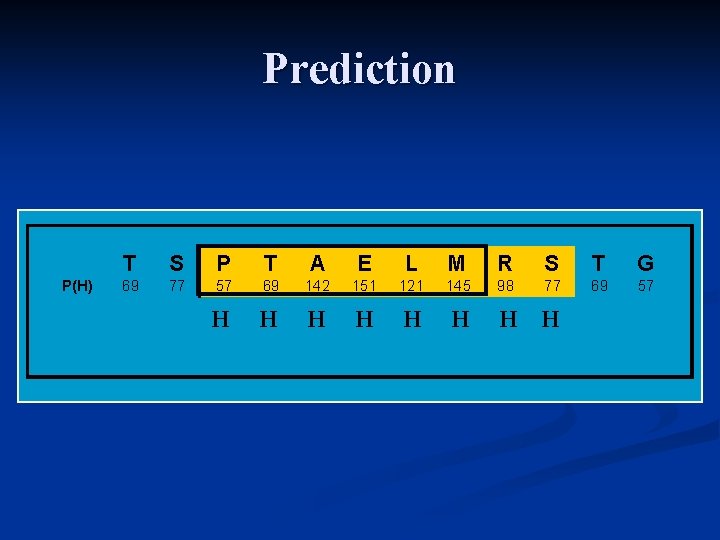
Prediction P(H) T S P T A E L M R S T G 69 77 57 69 142 151 121 145 98 77 69 57 H H H H
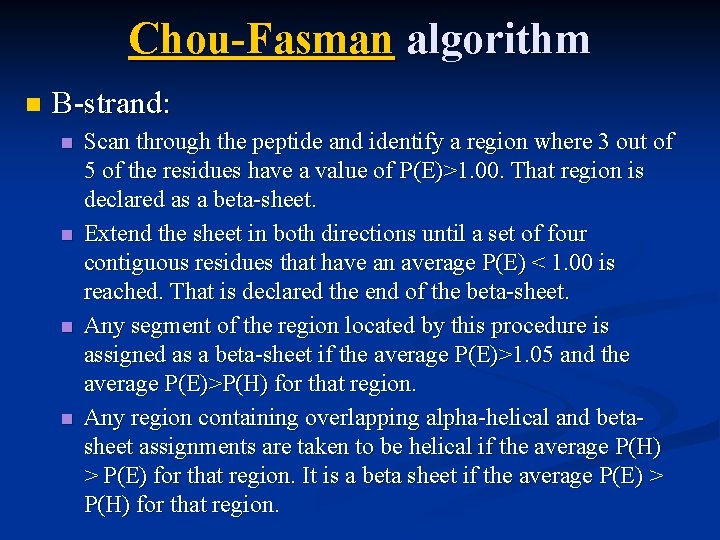
Chou-Fasman algorithm n B-strand: n n Scan through the peptide and identify a region where 3 out of 5 of the residues have a value of P(E)>1. 00. That region is declared as a beta-sheet. Extend the sheet in both directions until a set of four contiguous residues that have an average P(E) < 1. 00 is reached. That is declared the end of the beta-sheet. Any segment of the region located by this procedure is assigned as a beta-sheet if the average P(E)>1. 05 and the average P(E)>P(H) for that region. Any region containing overlapping alpha-helical and betasheet assignments are taken to be helical if the average P(H) > P(E) for that region. It is a beta sheet if the average P(E) > P(H) for that region.

Chou-Fasman algorithm n Beta-turn n To identify a bend at residue number j, calculate the following value p(t) = f(j)f(j+1)f(j+2)f(j+3) n If (1) (2) (3) p(t) > 0. 000075, the average value for P(turn) > 1. 00 in the tetrapeptide and the averages for the tetrapeptide obey the inequality P(H) < P(turn) > P(E), then a beta-turn is predicted at that location.

Exercise n Predict the secondary structure of the following protein sequence: Ala Pro Ala Phe Ser Val Ser Leu Ala Ser Gly Ala 142 57 142 113 77 106 77 121 142 77 57 142 83 55 83 138 75 170 75 130 83 75 75 83 66 152 66 60 143 59 66 143 156 66

exercise n Predict the secondary structure of the following protein sequence: Ala Pro Ala Phe Ser Val Ser Leu Ala Ser Gly Ala 142 H 83 57 H 55 113 H 138 E 60 77 H 75 E 143 106 H 170 E 50 77 121 142 77 57 142 75 E 143 130 E 59 T 83 E 66 T 75 83 66 142 H 83 E 152 66 143 156 66 T T T H H E E E T T E 75 T T
Prediction Methods n Single sequence n n Examine single protein sequence Base prediction on n Statistics – composition of amino acids Neural networks – patterns of amino acids Multiple sequence alignment n First create MSA n n Use sequences from PSI-BLAST, CLUSTALW, etc… Align sequence with related proteins in family Predict secondary structure based on consensus/profile Generally improves prediction 8 -9%
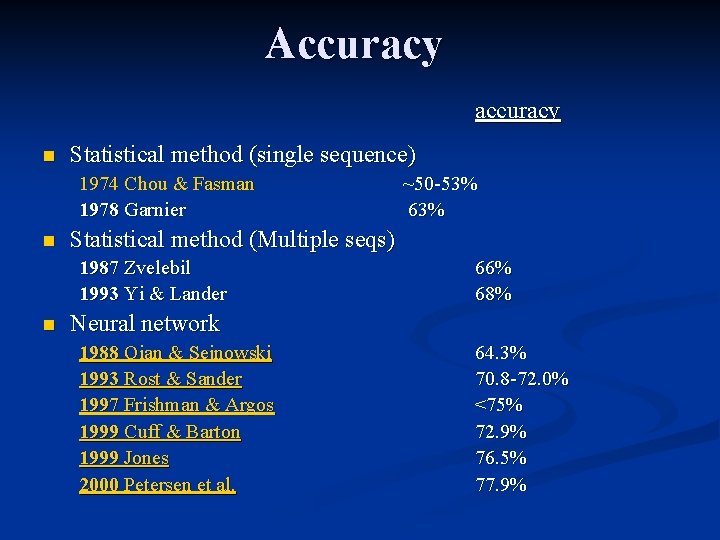
Accuracy accuracy n Statistical method (single sequence) 1974 Chou & Fasman 1978 Garnier n Statistical method (Multiple seqs) 1987 Zvelebil 1993 Yi & Lander n ~50 -53% 66% 68% Neural network 1988 Qian & Sejnowski 1993 Rost & Sander 1997 Frishman & Argos 1999 Cuff & Barton 1999 Jones 2000 Petersen et al. 64. 3% 70. 8 -72. 0% <75% 72. 9% 76. 5% 77. 9%
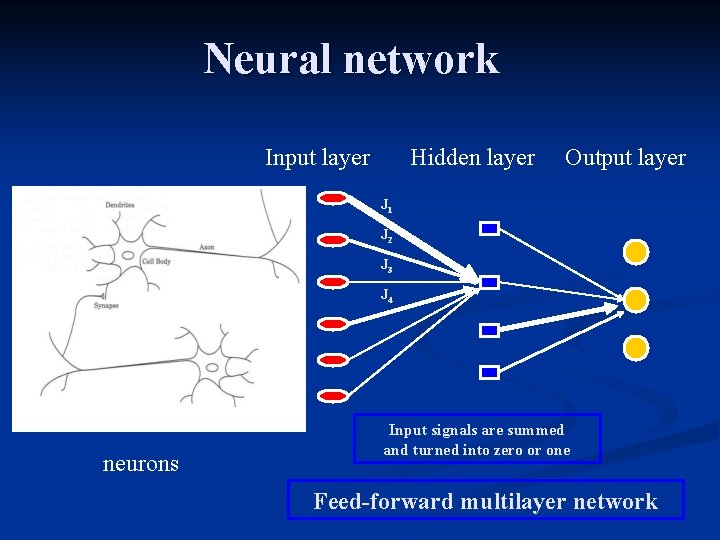
Neural network Input layer Hidden layer Output layer J 1 J 2 J 3 J 4 neurons Input signals are summed and turned into zero or one Feed-forward multilayer network
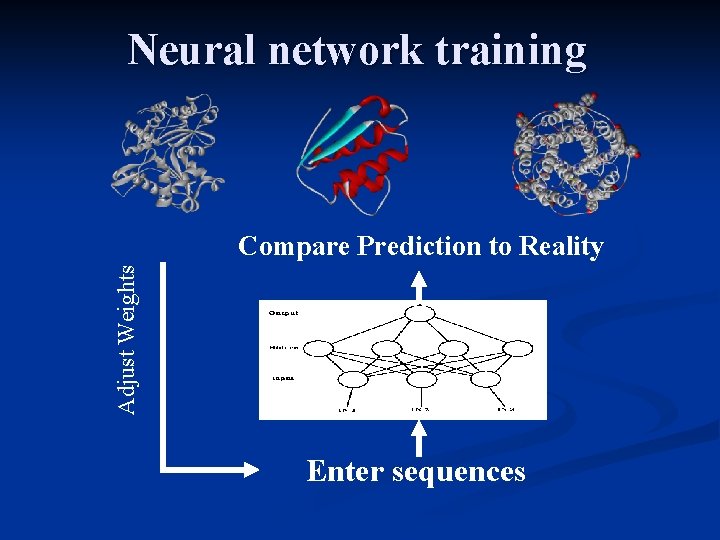
Neural network training Adjust Weights Compare Prediction to Reality Enter sequences

Neural net for secondary structure D (L) R (E) A C D E F G H I K L M N P Q R S T V W Y. Q (E) G (E) F (E) V (E) P (E) A (H) Y (H) V (E) K (E) H E L
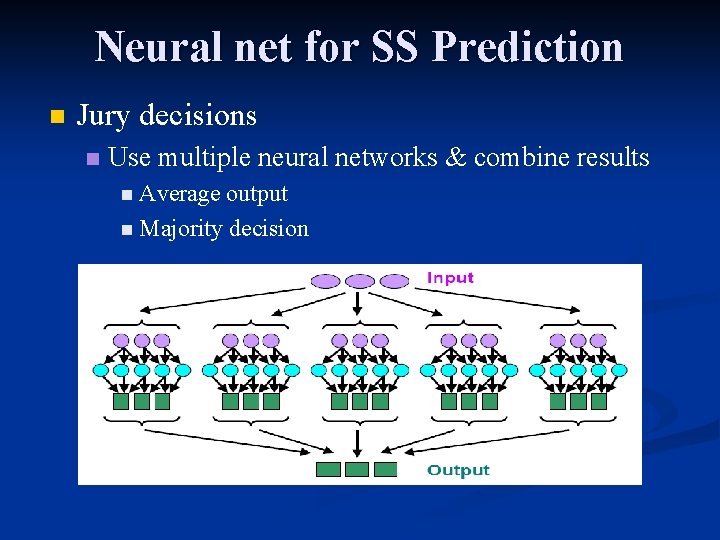
Neural net for SS Prediction n Jury decisions n Use multiple neural networks & combine results n Average output n Majority decision
![Neural net for SS Prediction n JPRED [Cuff+ 1998] n Finds consensus from PHD, Neural net for SS Prediction n JPRED [Cuff+ 1998] n Finds consensus from PHD,](http://slidetodoc.com/presentation_image_h/bd50bc77189437fe7b122a941e740169/image-40.jpg)
Neural net for SS Prediction n JPRED [Cuff+ 1998] n Finds consensus from PHD, PREDATOR, DSC, NNSSP, etc…
- Slides: 40