Protein Structure Alignment Human Hemoglobin alphachain pdb 1
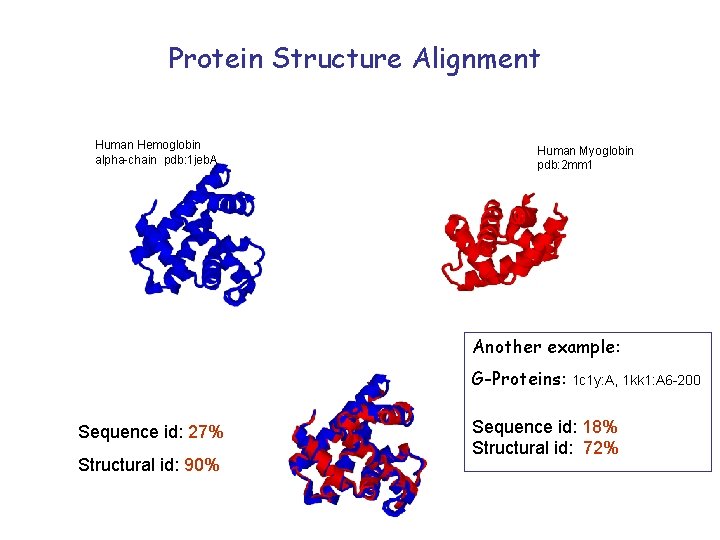
Protein Structure Alignment Human Hemoglobin alpha-chain pdb: 1 jeb. A Human Myoglobin pdb: 2 mm 1 Another example: G-Proteins: Sequence id: 27% Structural id: 90% 1 c 1 y: A, 1 kk 1: A 6 -200 Sequence id: 18% Structural id: 72%
Transformations Translation and Rotation n Rigid Motion (Euclidian Trans. ) Translation, Rotation + Scaling
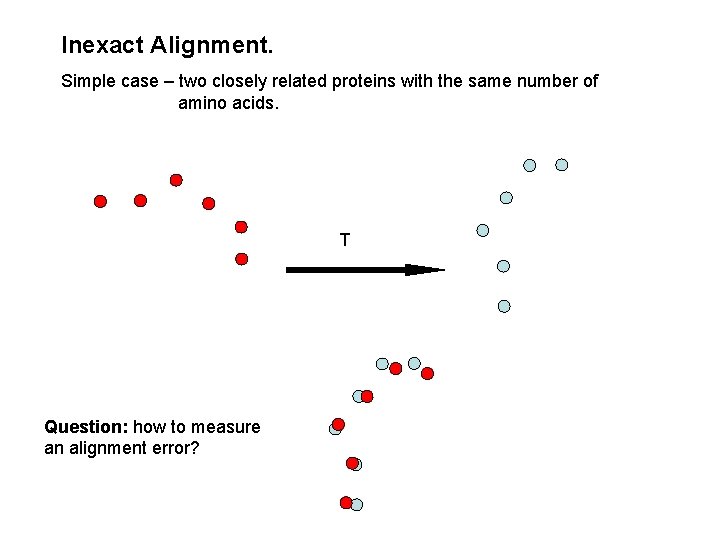
Inexact Alignment. Simple case – two closely related proteins with the same number of amino acids. T Question: how to measure an alignment error?

Distance Functions Two point sets: A={ai} i=1…n B={bj} j=1…m • Pairwise Correspondence: (ak 1, bt 1) (ak 2, bt 2)… (ak. N, bt. N) (1) Exact Matching: ||aki – bti||=0 (2) Bottleneck max ||aki – bti|| (3) RMSD (Root Mean Square Distance) Sqrt( Σ||aki – bti||2/N)
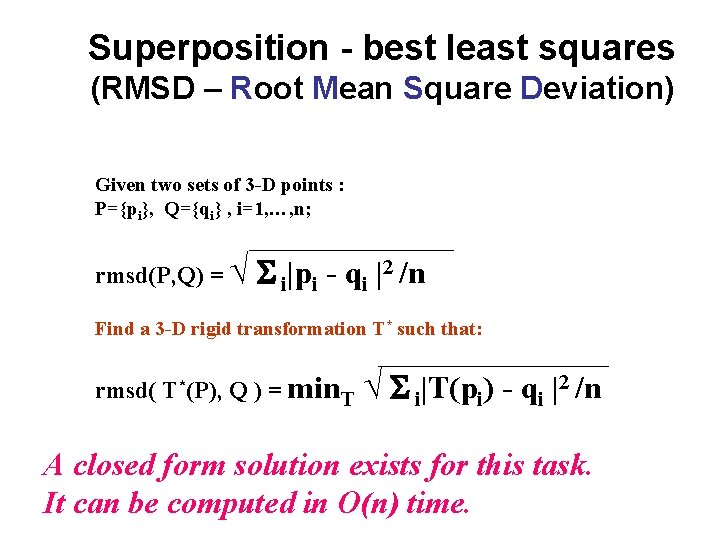
Superposition - best least squares (RMSD – Root Mean Square Deviation) Given two sets of 3 -D points : P={pi}, Q={qi} , i=1, …, n; rmsd(P, Q) = √ S i|pi - qi |2 /n Find a 3 -D rigid transformation T* such that: rmsd( T*(P), Q ) = min. T √ S i|T(pi) - qi |2 /n A closed form solution exists for this task. It can be computed in O(n) time.
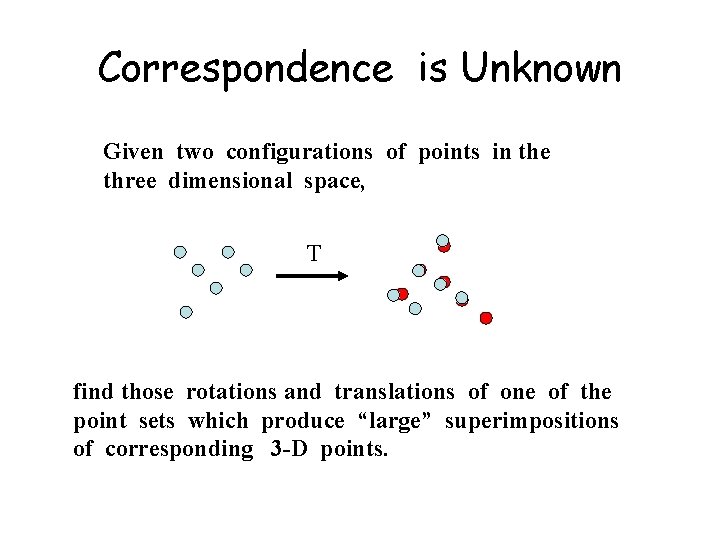
Correspondence is Unknown Given two configurations of points in the three dimensional space, T find those rotations and translations of one of the point sets which produce “large” superimpositions of corresponding 3 -D points.
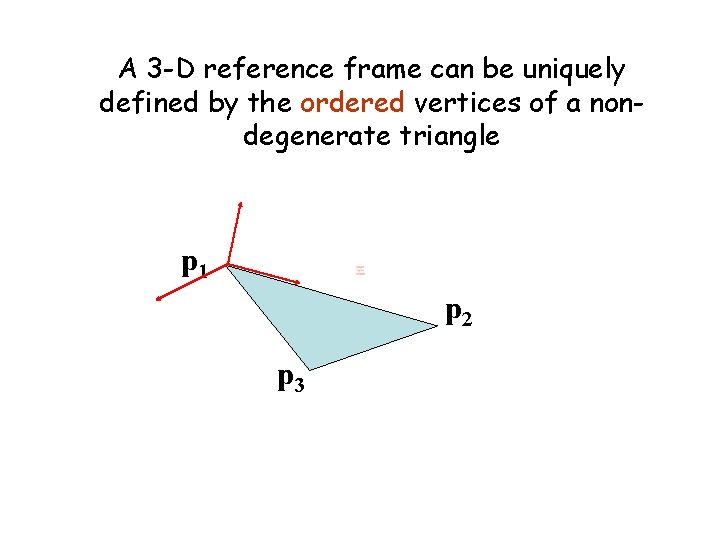
A 3 -D reference frame can be uniquely defined by the ordered vertices of a nondegenerate triangle p 1 p 2 p 3
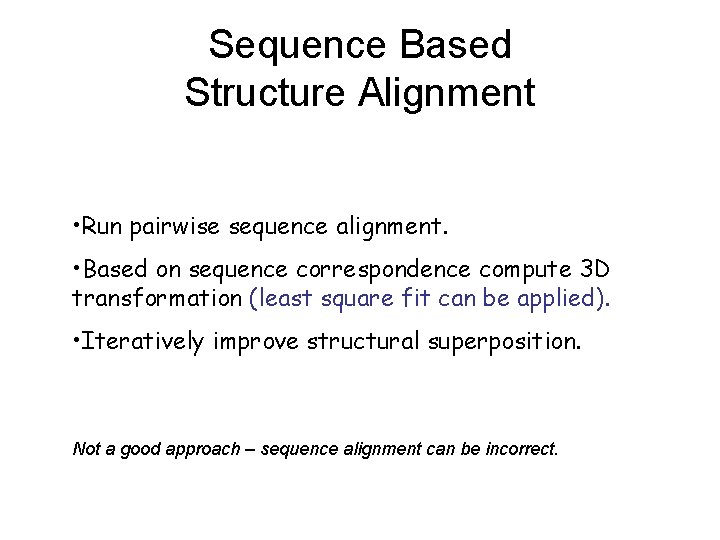
Sequence Based Structure Alignment • Run pairwise sequence alignment. • Based on sequence correspondence compute 3 D transformation (least square fit can be applied). • Iteratively improve structural superposition. Not a good approach – sequence alignment can be incorrect.
Structure Alignment (Straightforward Algorithm) • For each pair of triplets, one from each molecule which define ‘almost’ congruent triangles compute the rigid transformation that superimposes them. • Count the number of aligned point pairs and sort the hypotheses by this number.
• For the highest ranking hypotheses improve the transformation by replacing it by the best RMSD transformation for all the matching pairs. • Complexity : O(n 3 m 3 ) * O(nm). Applying 3 D grid gives practically O(n 3 m 3) * O(n) • If one exploits protein backbone geometry + 3 D grid : O(nm) * O(n)
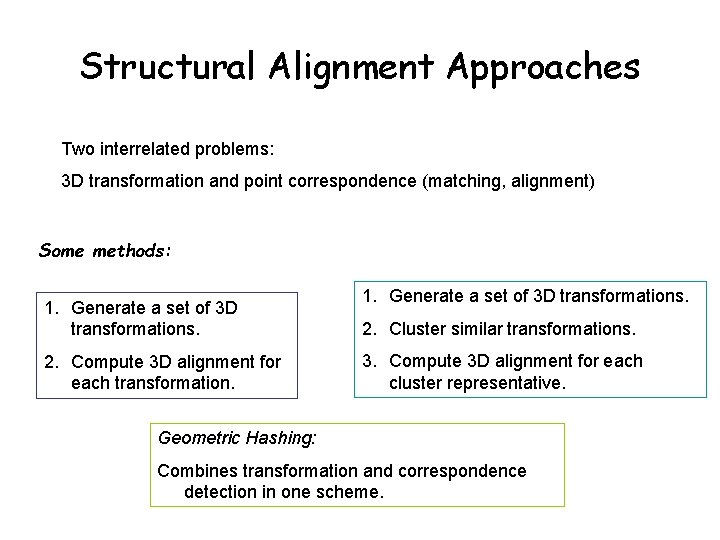
Structural Alignment Approaches Two interrelated problems: 3 D transformation and point correspondence (matching, alignment) Some methods: 1. Generate a set of 3 D transformations. 2. Compute 3 D alignment for each transformation. 1. Generate a set of 3 D transformations. 2. Cluster similar transformations. 3. Compute 3 D alignment for each cluster representative. Geometric Hashing: Combines transformation and correspondence detection in one scheme.
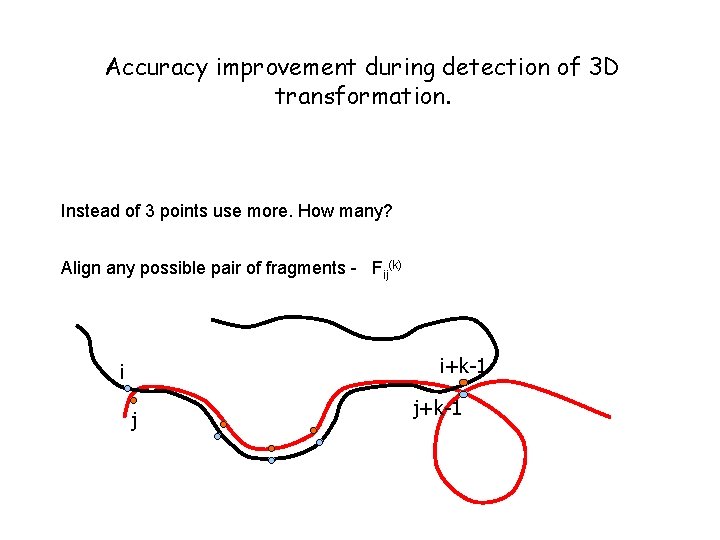
Accuracy improvement during detection of 3 D transformation. Instead of 3 points use more. How many? Align any possible pair of fragments - Fij(k) i+k-1 i j j+k-1

Accept Fij(k) if rmsd(Fij(k)) <e. Complexity O(n 3 n) * O(n) (For each Fij(k) we (assume n~m) need compute its rmsd) can be reduced to O(n 3) * O(n)
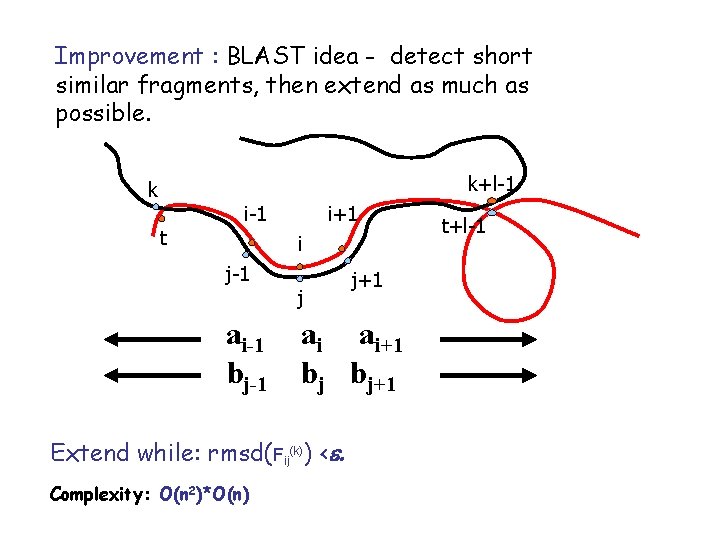
Improvement : BLAST idea - detect short similar fragments, then extend as much as possible. k+l-1 k t i-1 i+1 i j-1 ai-1 bj-1 j ai ai+1 bj bj+1 Extend while: rmsd(Fij(k)) <e. Complexity: O(n 2)*O(n) j+1 t+l-1
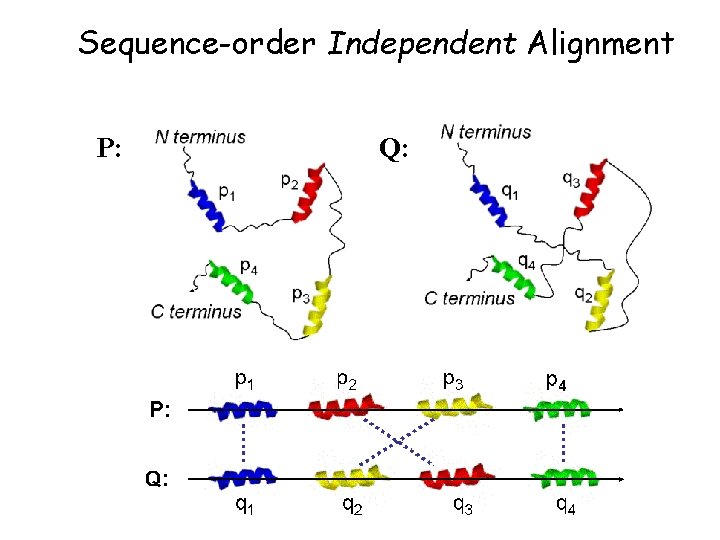
Sequence-order Independent Alignment P: Q:
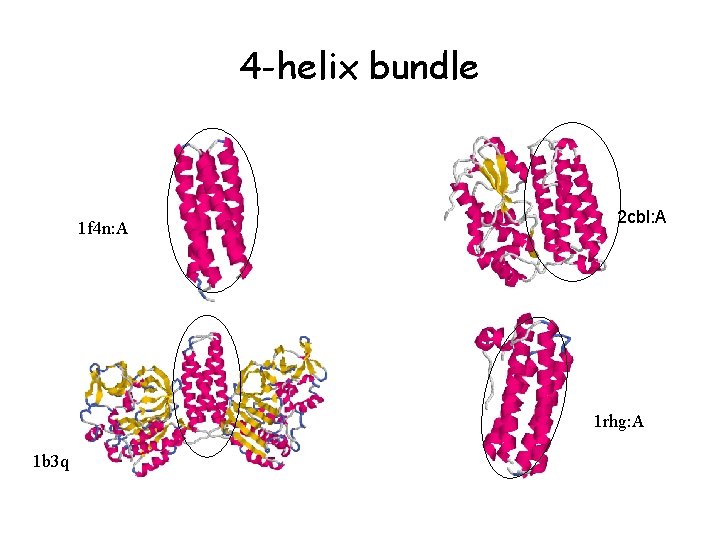
4 -helix bundle 1 f 4 n: A 2 cbl: A 1 rhg: A 1 b 3 q
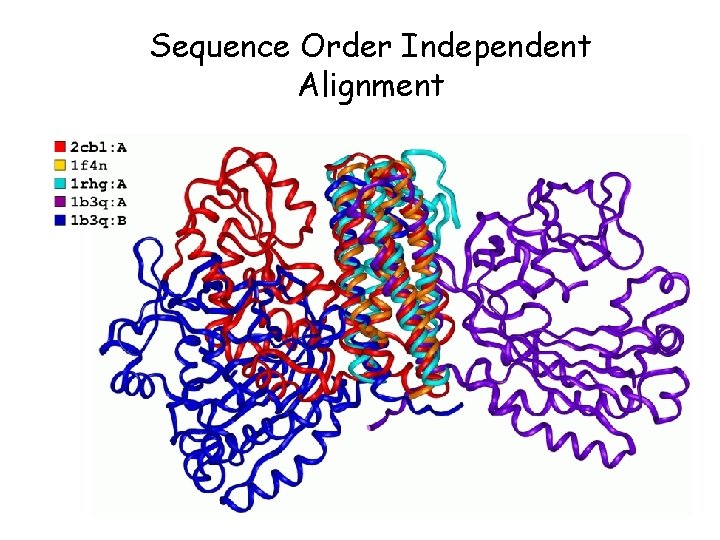
Sequence Order Independent Alignment
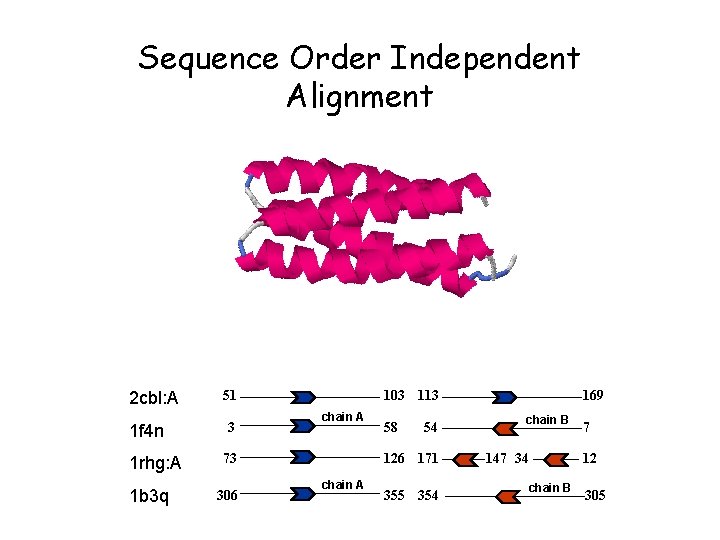
Sequence Order Independent Alignment 2 cbl: A 1 f 4 n 1 rhg: A 1 b 3 q 51 3 103 113 chain A 73 306 58 54 126 171 chain A 355 354 169 chain B 147 34 7 12 chain B 305
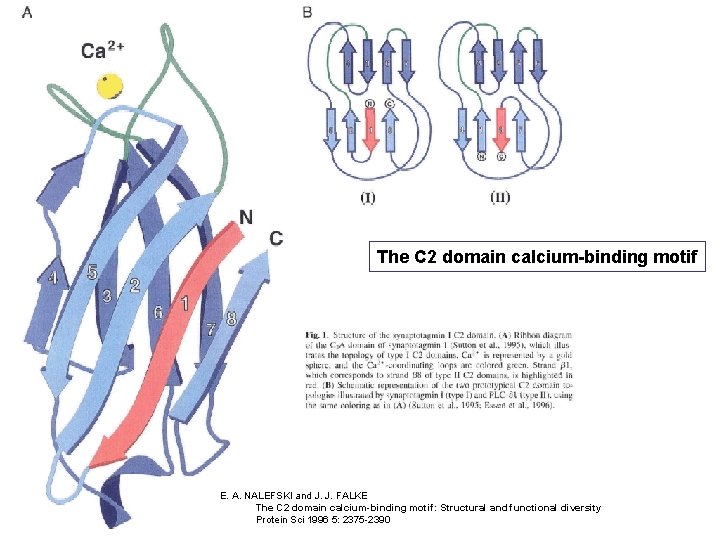
The C 2 domain calcium-binding motif E. A. NALEFSKI and J. J. FALKE The C 2 domain calcium-binding motif: Structural and functional diversity Protein Sci 1996 5: 2375 -2390
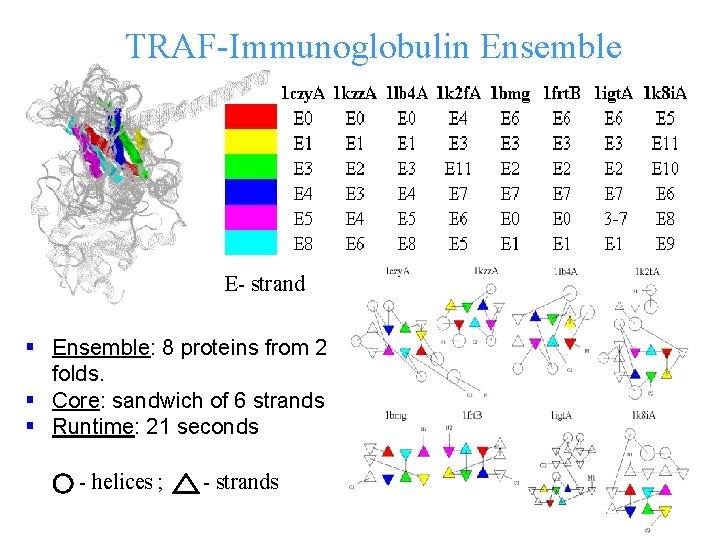
TRAF-Immunoglobulin Ensemble E- strand § Ensemble: 8 proteins from 2 folds. § Core: sandwich of 6 strands § Runtime: 21 seconds - helices ; - strands
Some Links • Rasmol – Molecular Visualization • SCOP - Structural Classification of Proteins • Multi. Prot - Protein Structural (pairwise/multiple) Alignment • MASS – Secondary Structure Based (pairwise/multiple) Alignment
- Slides: 21