Protein Structure 1 Primary and Secondary Structure Proteins
- Slides: 15

Protein Structure 1 Primary and Secondary Structure

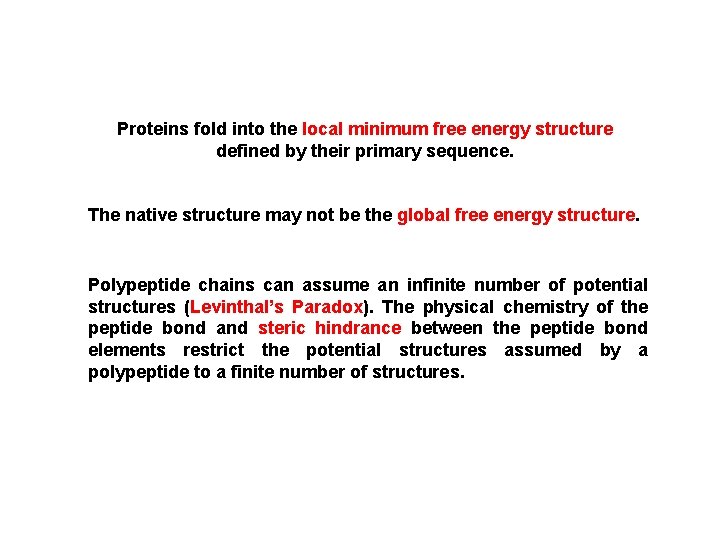
Proteins fold into the local minimum free energy structure defined by their primary sequence. The native structure may not be the global free energy structure. Polypeptide chains can assume an infinite number of potential structures (Levinthal’s Paradox). The physical chemistry of the peptide bond and steric hindrance between the peptide bond elements restrict the potential structures assumed by a polypeptide to a finite number of structures.

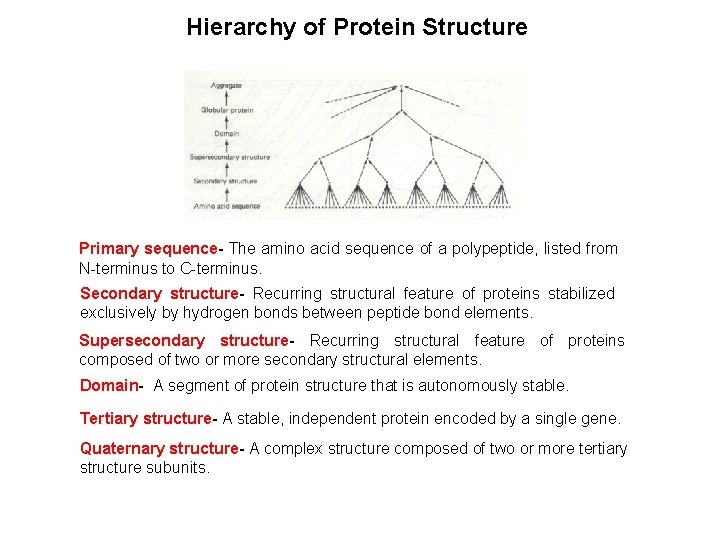
Hierarchy of Protein Structure Primary sequence The amino acid sequence of a polypeptide, listed from N-terminus to C-terminus. Secondary structure Recurring structural feature of proteins stabilized exclusively by hydrogen bonds between peptide bond elements. Supersecondary structure Recurring structural feature of proteins composed of two or more secondary structural elements. Domain- A segment of protein structure that is autonomously stable. Tertiary structure A stable, independent protein encoded by a single gene. Quaternary structure A complex structure composed of two or more tertiary structure subunits.

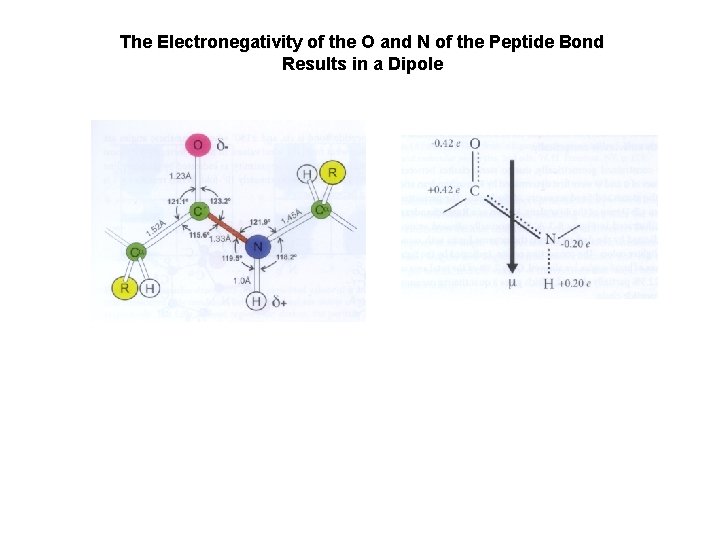
The Electronegativity of the O and N of the Peptide Bond Results in a Dipole

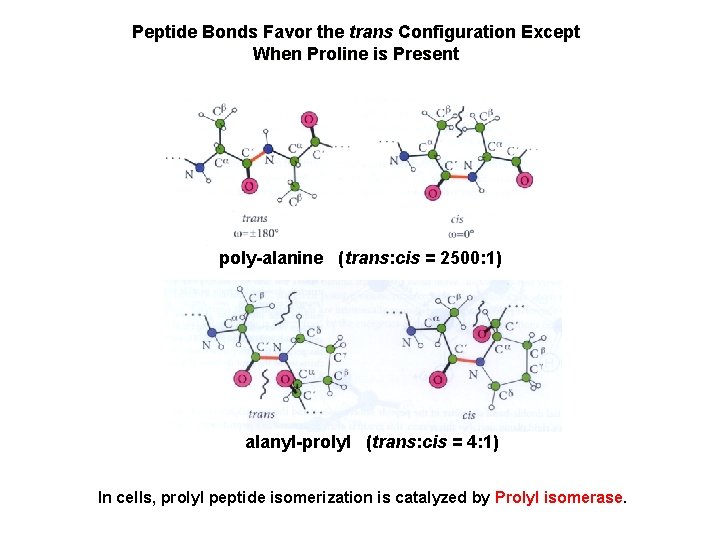
Peptide Bonds Favor the trans Configuration Except When Proline is Present poly-alanine (trans: cis = 2500: 1) alanyl-prolyl (trans: cis = 4: 1) In cells, prolyl peptide isomerization is catalyzed by Prolyl isomerase.

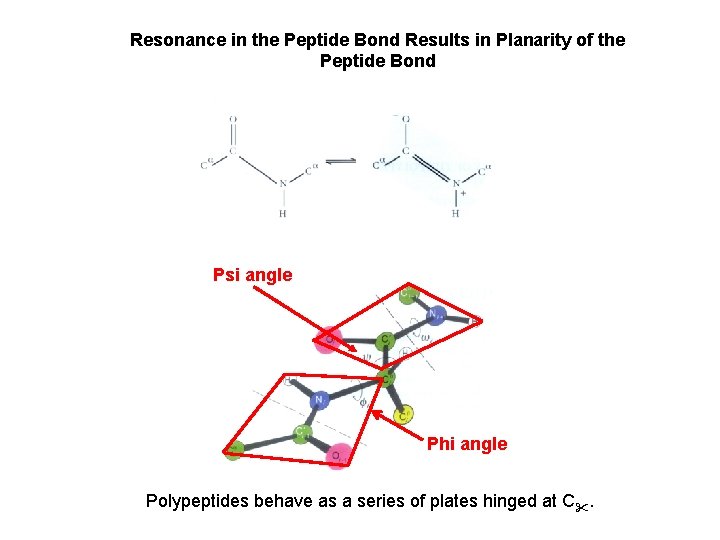
Resonance in the Peptide Bond Results in Planarity of the Peptide Bond Psi angle Phi angle Polypeptides behave as a series of plates hinged at C.

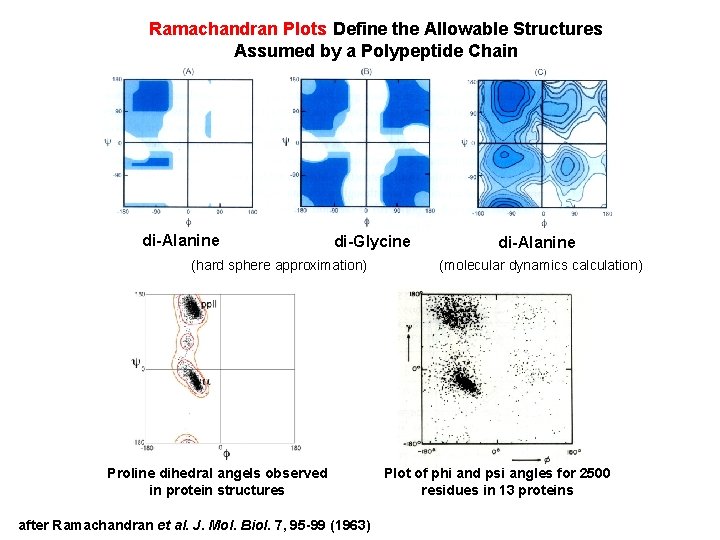
Ramachandran Plots Define the Allowable Structures Assumed by a Polypeptide Chain di-Alanine di-Glycine (hard sphere approximation) Proline dihedral angels observed in protein structures after Ramachandran et al. J. Mol. Biol. 7, 95 -99 (1963) di-Alanine (molecular dynamics calculation) Plot of phi and psi angles for 2500 residues in 13 proteins

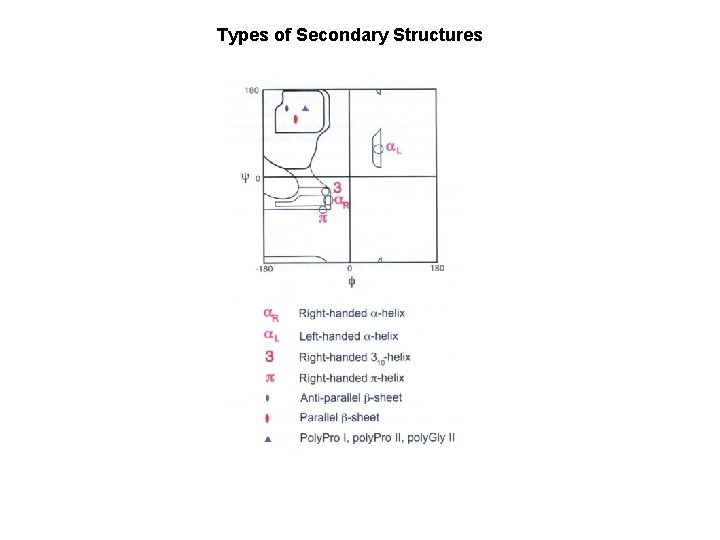
Types of Secondary Structures

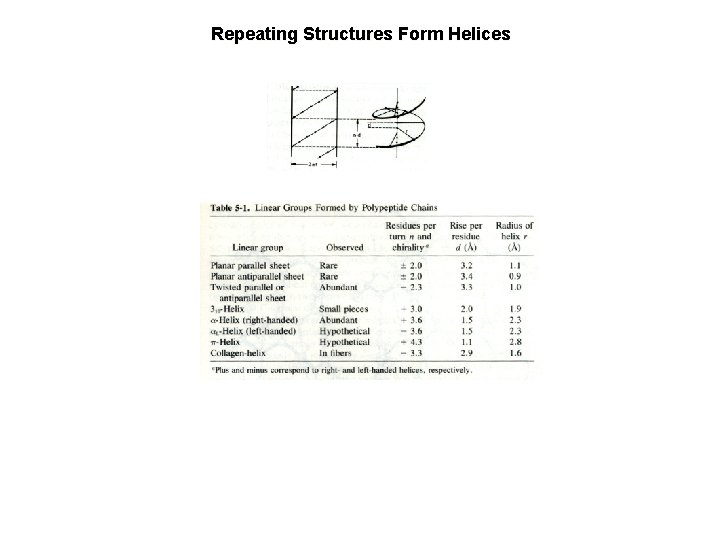
Repeating Structures Form Helices

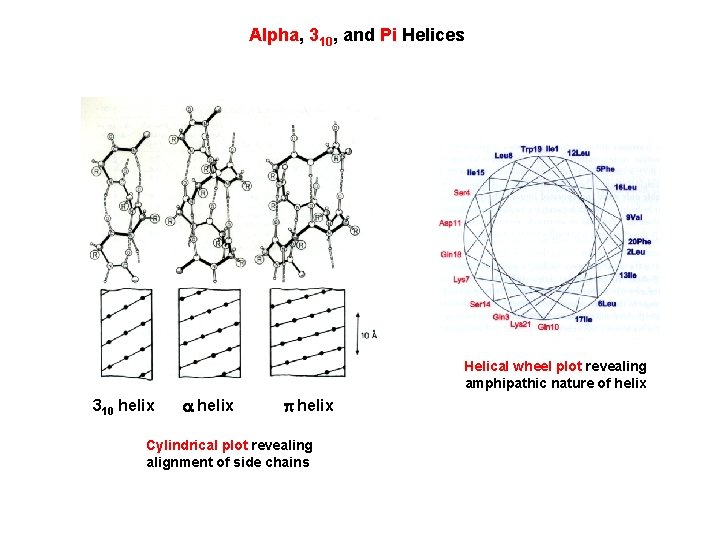
Alpha, 310, and Pi Helices Helical wheel plot revealing amphipathic nature of helix 310 helix Cylindrical plot revealing alignment of side chains

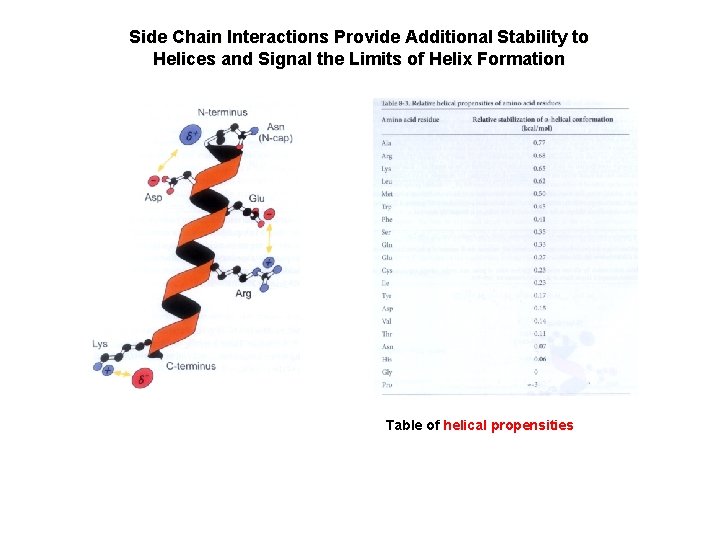
Side Chain Interactions Provide Additional Stability to Helices and Signal the Limits of Helix Formation Table of helical propensities

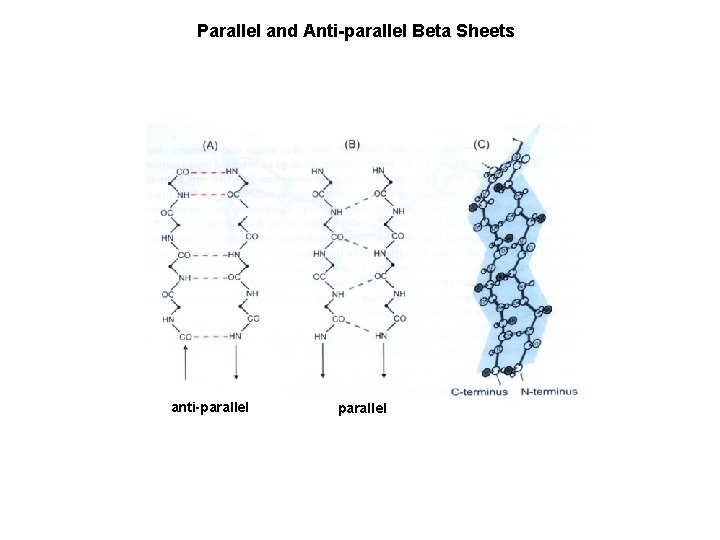
Parallel and Anti-parallel Beta Sheets anti-parallel

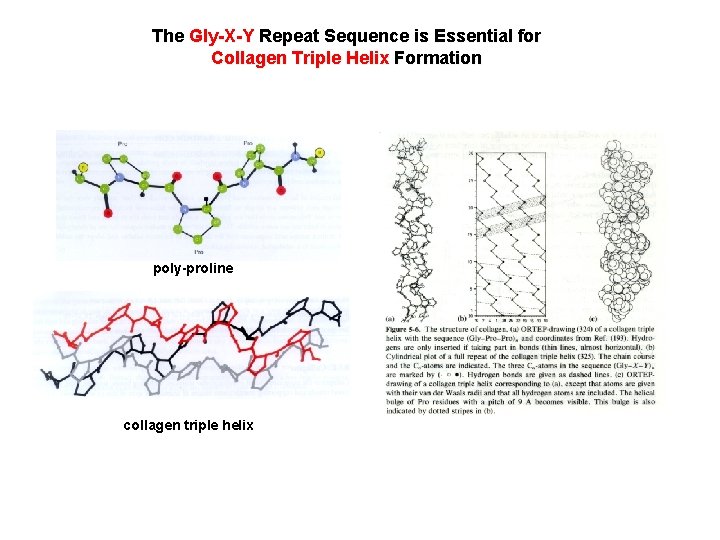
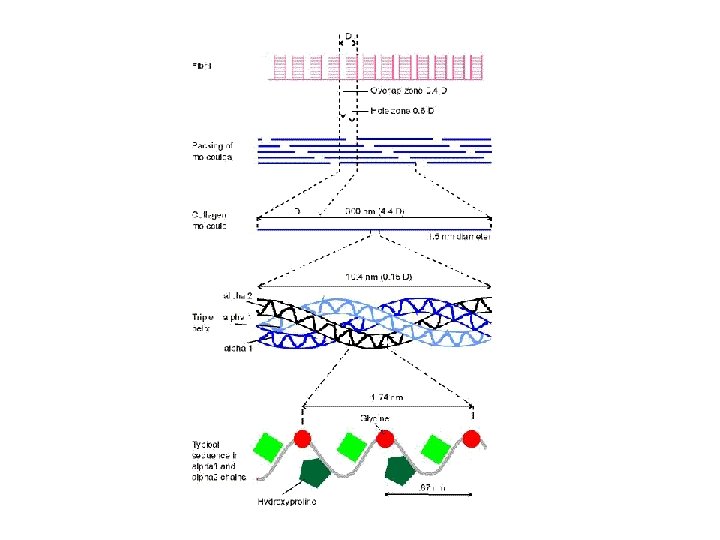
The Gly-X-Y Repeat Sequence is Essential for Collagen Triple Helix Formation poly-proline collagen triple helix


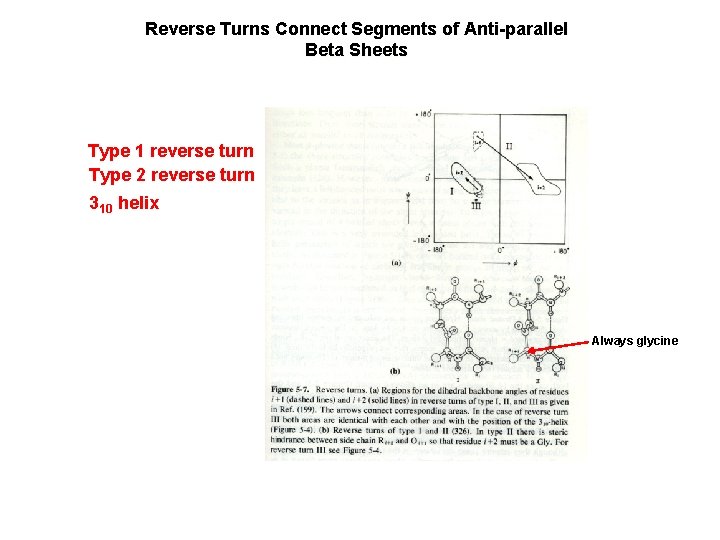
Reverse Turns Connect Segments of Anti-parallel Beta Sheets Type 1 reverse turn Type 2 reverse turn 310 helix Always glycine