PROTEIN SEQUENCING PRESENTATION OUTLINE INTRODUCTION PROTEIN SEQUENCING WHY




- Slides: 29

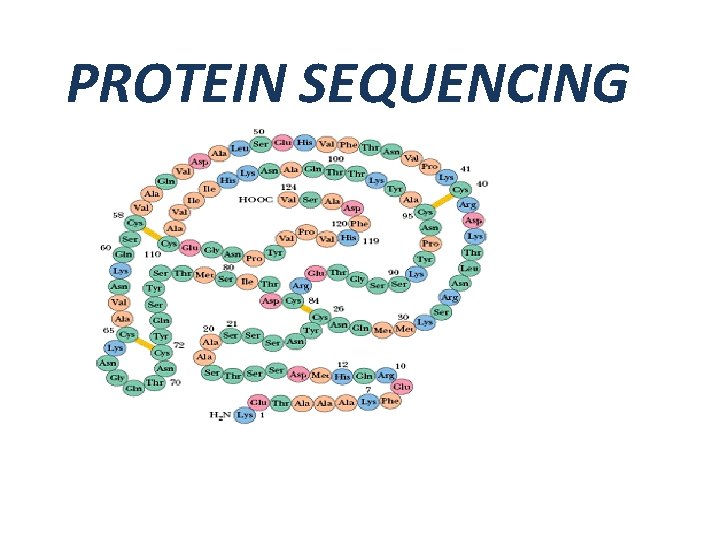
PROTEIN SEQUENCING

PRESENTATION OUTLINE • • INTRODUCTION PROTEIN SEQUENCING WHY PROTEIN SEQUENCING MATTERS? HISTORY STRATEGY SEQUENCING METHODS APPLICATIONS

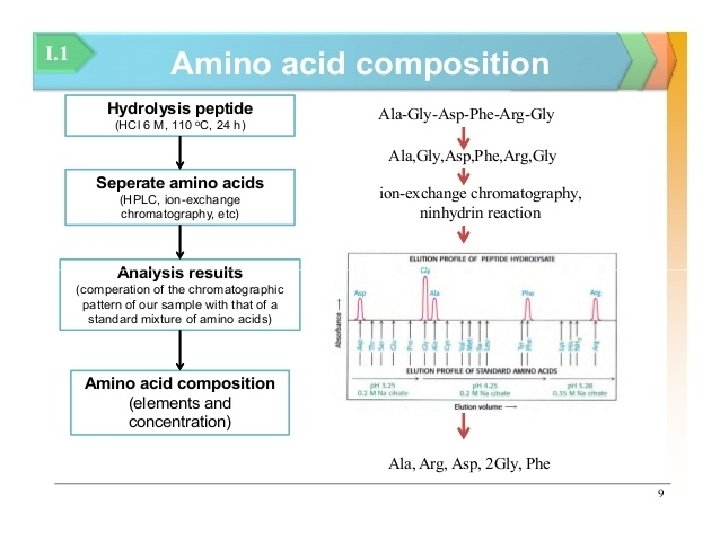
INTRODUCTION q PROTEIN • Proteins are polymers of amino acid • Protein's structure & function depends upon amino acid sequence q. PROTEIN SEQUENCING • Technique to find out the amino acid sequence • Important for understanding cellular process

WHY DOES SEQUENCE MATTER? • Sequences and composition often reflect the function of the protein(often proteins of similar function will have similar sequences). • Homologous protein from different organisms have homologous sequences. • Changes in protein sequence can be used to infer evolutionary relationships.

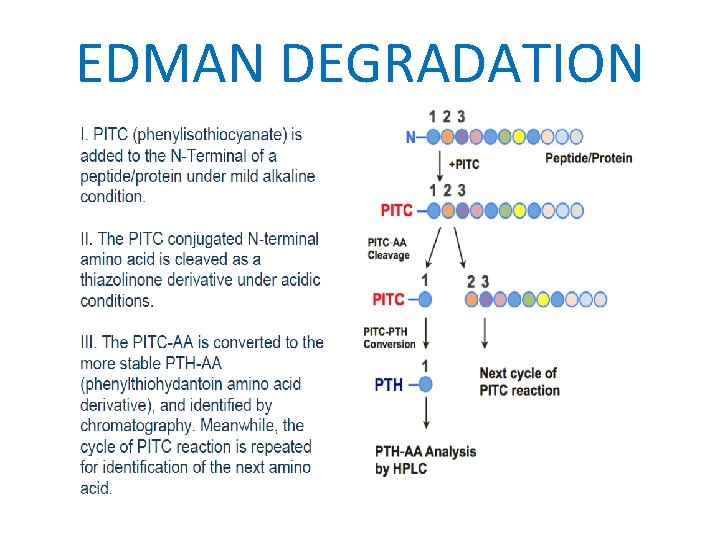
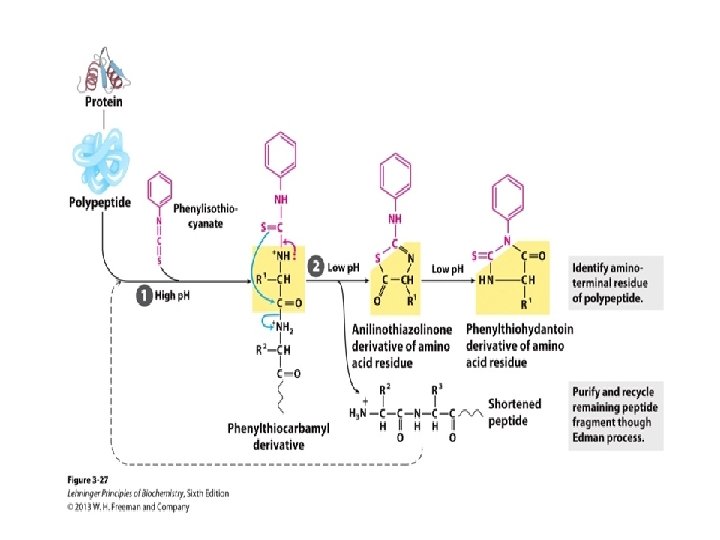
HISTORY • Pehr Edman 1950 He found the method to decode the amino acid sequence of protein using chemicals. • Fredrick Sanger 1955 He was able to present the complete sequence of INSULIN.


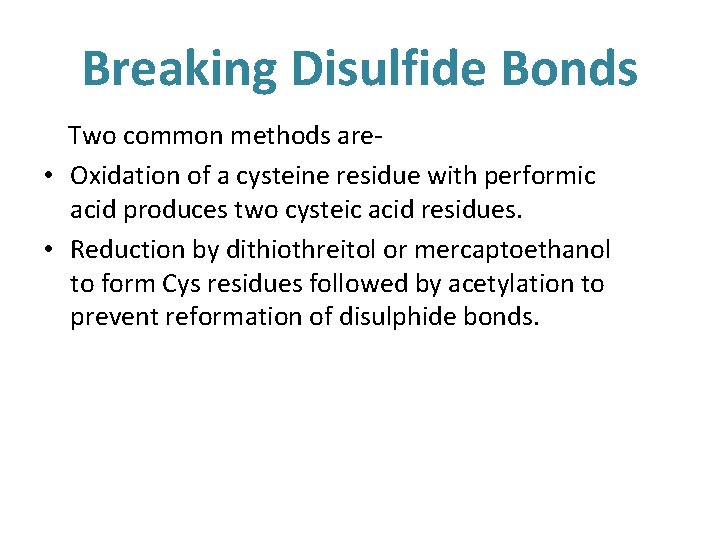
SEPARATION OF CHAINS • Subunits interaction depend on weak forces • Separation of polypeptide chains is achieved with – ü Extreme ph ü Urea ü Guanidine HCl

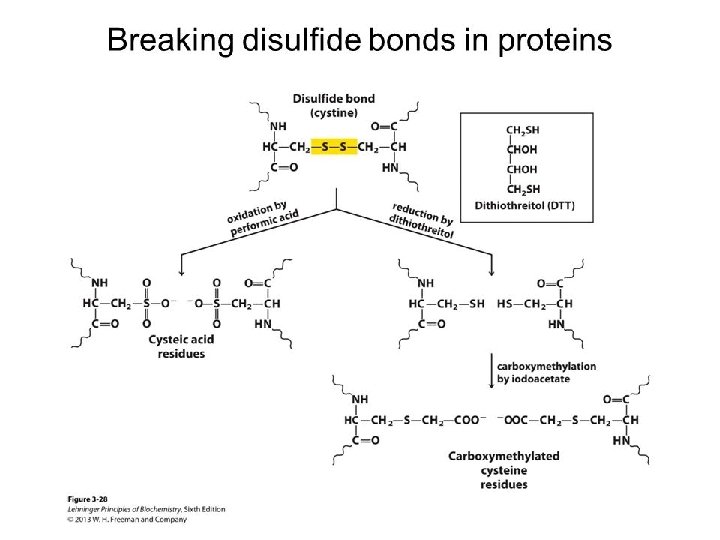
Breaking Disulfide Bonds Two common methods are • Oxidation of a cysteine residue with performic acid produces two cysteic acid residues. • Reduction by dithiothreitol or mercaptoethanol to form Cys residues followed by acetylation to prevent reformation of disulphide bonds.




z

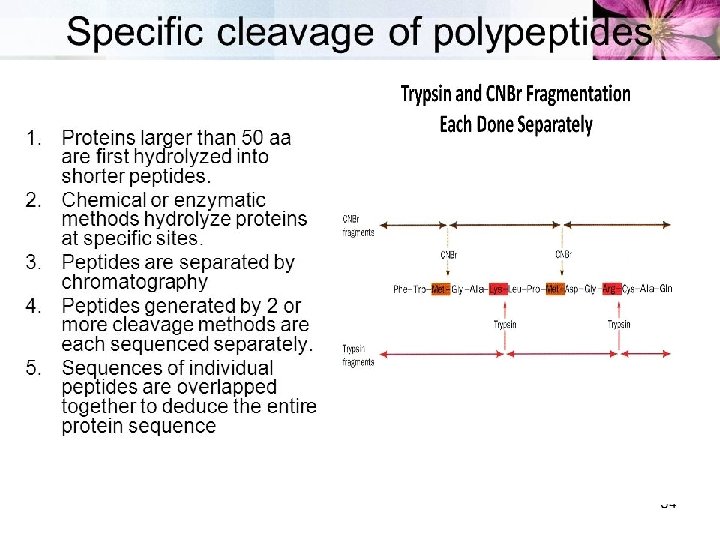
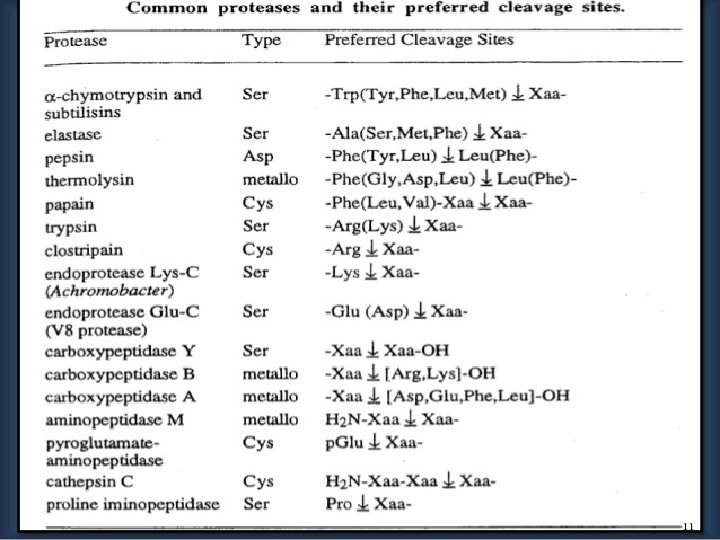
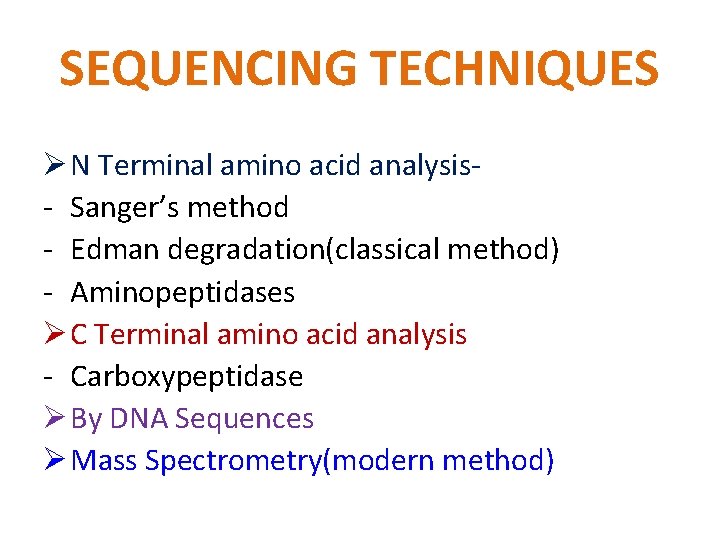
SEQUENCING TECHNIQUES Ø N Terminal amino acid analysis- Sanger’s method - Edman degradation(classical method) - Aminopeptidases Ø C Terminal amino acid analysis - Carboxypeptidase Ø By DNA Sequences Ø Mass Spectrometry(modern method)

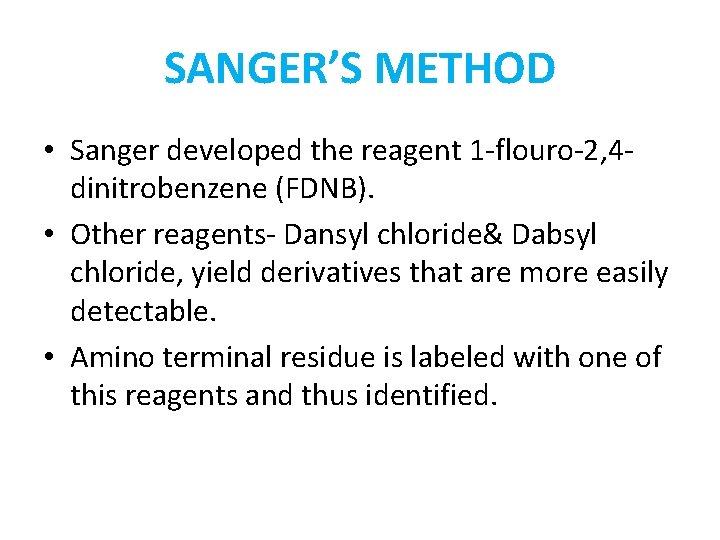
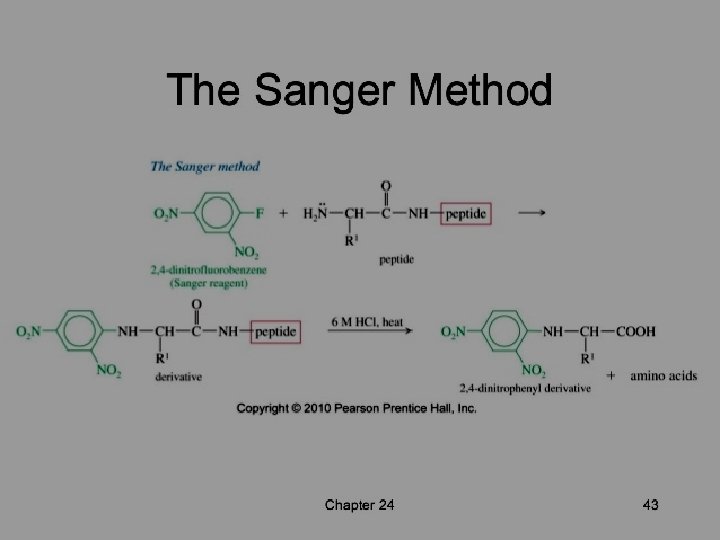
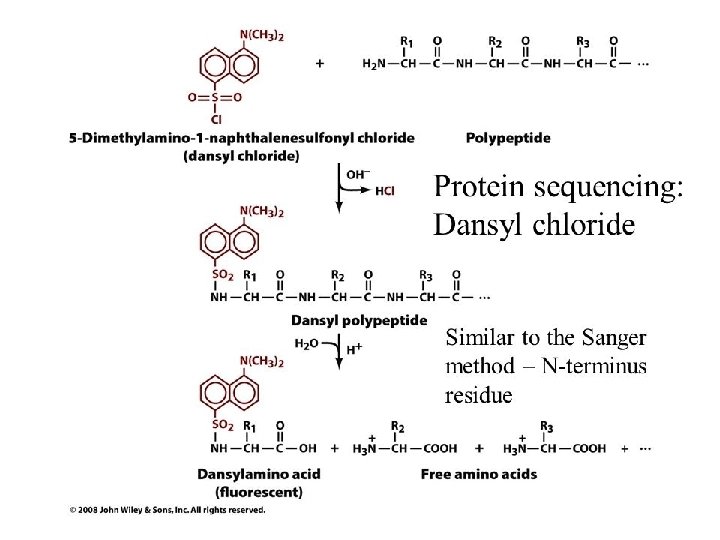
SANGER’S METHOD • Sanger developed the reagent 1 -flouro-2, 4 dinitrobenzene (FDNB). • Other reagents- Dansyl chloride& Dabsyl chloride, yield derivatives that are more easily detectable. • Amino terminal residue is labeled with one of this reagents and thus identified.

• Advantageh - It can help determine the no of chemically distinct polypeptides in a protein, provided each has a different amino terminal residue. • Disadvantage - Hydrolysis stage destroys polypeptide beyond amino terminal residue.



EDMAN DEGRADATION



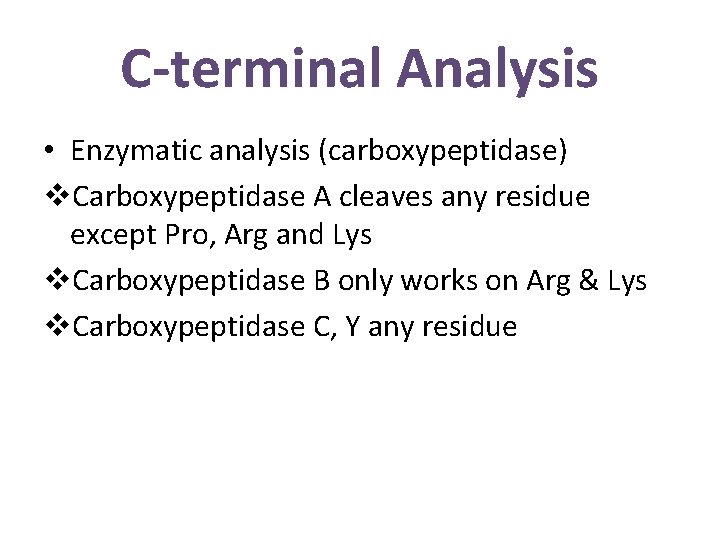
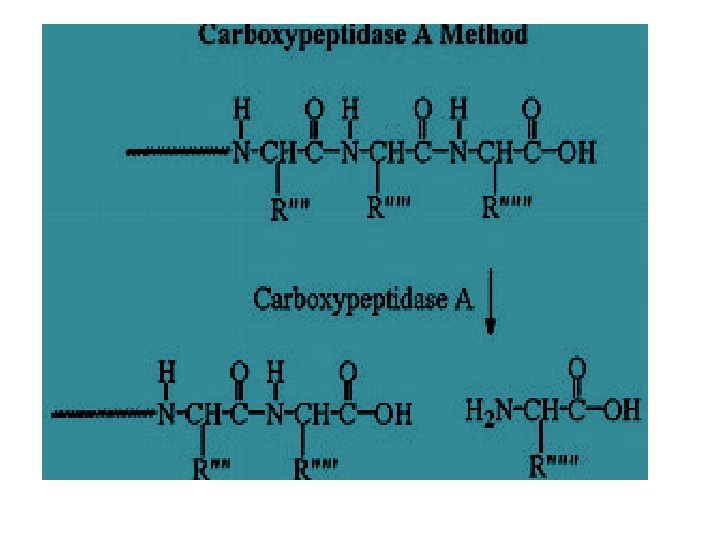
C-terminal Analysis • Enzymatic analysis (carboxypeptidase) v. Carboxypeptidase A cleaves any residue except Pro, Arg and Lys v. Carboxypeptidase B only works on Arg & Lys v. Carboxypeptidase C, Y any residue



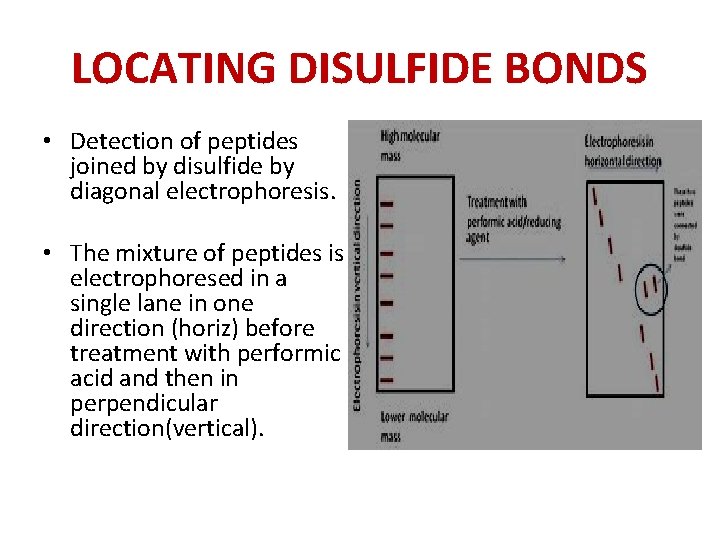
LOCATING DISULFIDE BONDS • Detection of peptides joined by disulfide by diagonal electrophoresis. • The mixture of peptides is electrophoresed in a single lane in one direction (horiz) before treatment with performic acid and then in perpendicular direction(vertical).

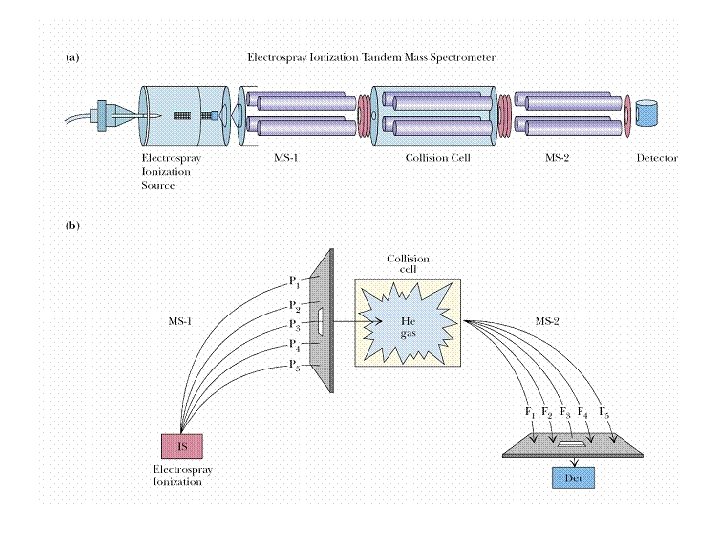
Protein Sequencing by Mass Spectrometry • Mass spectrometry(MS) is an analytical technique that measures the mass to charge ratio of charge particles. • MALDI MS and ESI MS can precisely identify the mass of a peptide and thus the amino acid sequence.


Prediction from DNA Sequence Protein sequence can also be determined indirectly from the DNA by determining nucleotide sequence in a gene. v Sequence a short section , perhaps 15 amino acids long of the protein. v Design primers from the amino acid sequence and amplify the gene , sequence the gene and determine the amino acid sequence.

APPLICATIONS 1) Comparison of protein sequences to establish similarities that define protein families. 2) Comparison of the same protein in different species to reveal evolutionary relationships. 3) Amino acid sequences can be searched for the presence of internal repeats. 4) Amino acid sequences contain signals that determine the destination of proteins and control their processing. 5) Sequence data provide a basis for preparing antibodies specific for a protein of interest.

THANK YOU