Protein Sequencing and Identification by Mass Spectrometry Masses

Protein Sequencing and Identification by Mass Spectrometry
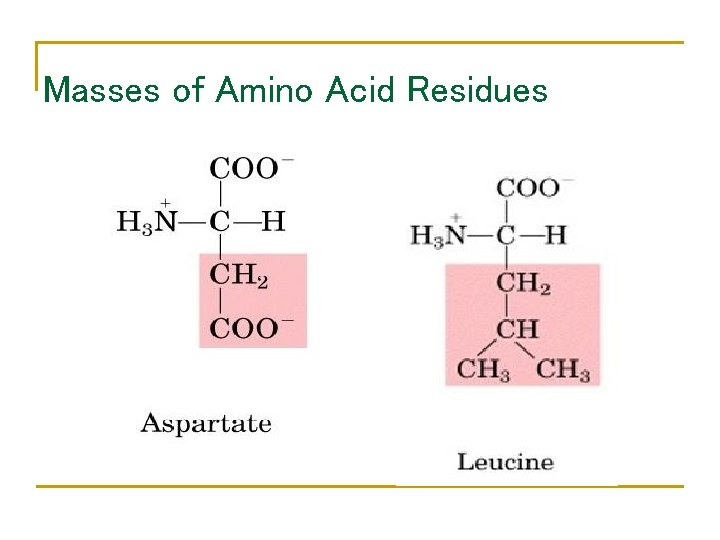
Masses of Amino Acid Residues
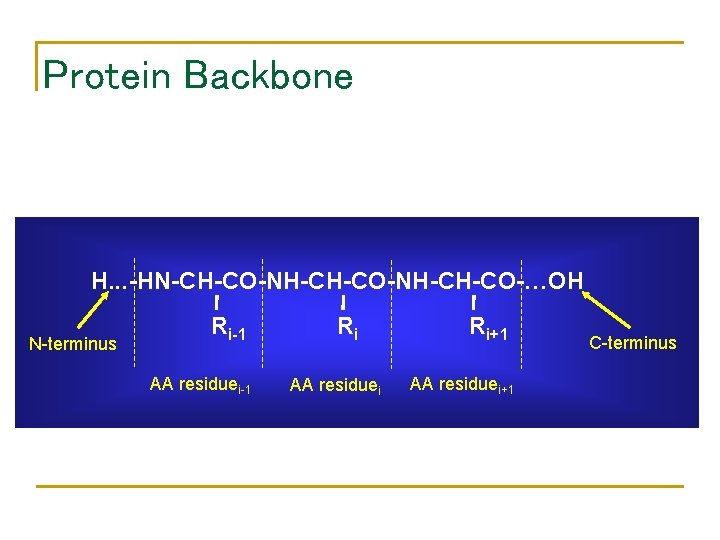
Protein Backbone H. . . -HN-CH-CO-NH-CH-CO-…OH N-terminus Ri-1 AA residuei-1 Ri AA residuei Ri+1 AA residuei+1 C-terminus

Peptide Fragmentation Collision Induced Dissociation H+ H. . . -HN-CH-CO Ri-1 Prefix Fragment • • . . . NH-CH-CO-…OH Ri Ri+1 Suffix Fragment Peptides tend to fragment along the backbone. Fragments can also loose neutral chemical groups like NH 3 and H 2 O.
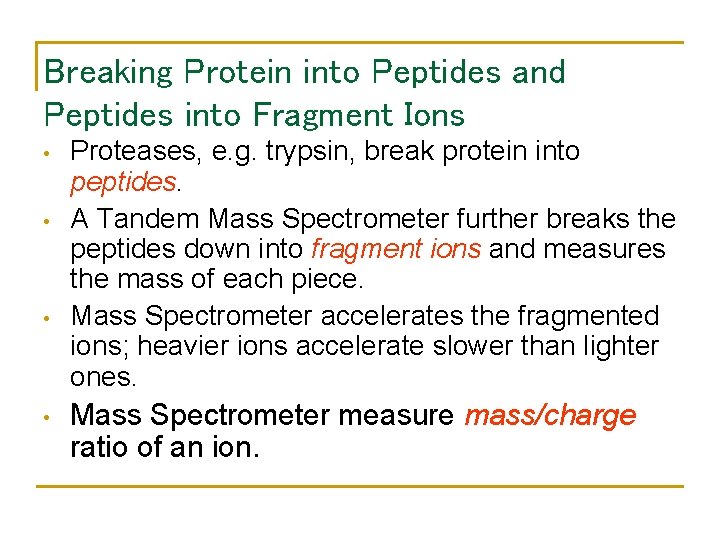
Breaking Protein into Peptides and Peptides into Fragment Ions • • Proteases, e. g. trypsin, break protein into peptides. A Tandem Mass Spectrometer further breaks the peptides down into fragment ions and measures the mass of each piece. Mass Spectrometer accelerates the fragmented ions; heavier ions accelerate slower than lighter ones. Mass Spectrometer measure mass/charge ratio of an ion.
rm te C- es tid ep es id pt pe lp ina al in rm te N- N- and C-terminal Peptides
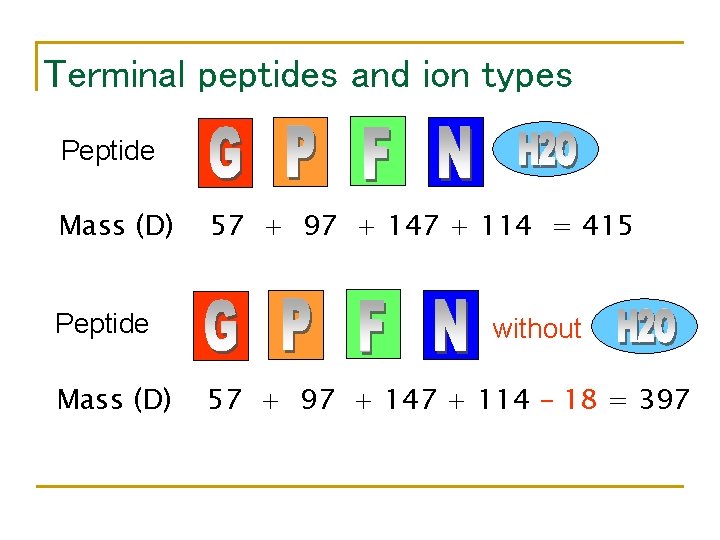
Terminal peptides and ion types Peptide Mass (D) 57 + 97 + 147 + 114 = 415 without 57 + 97 + 147 + 114 – 18 = 397
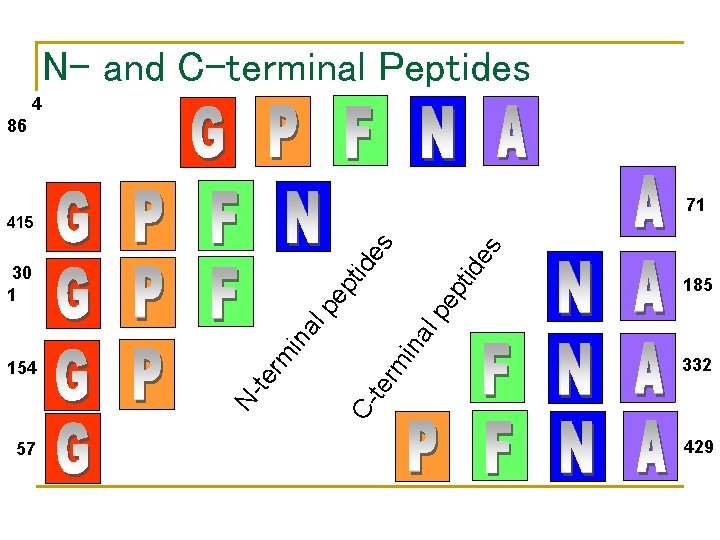
N- and C-terminal Peptides 4 86 71 ep pt 57 185 lp pe ina al 332 C- te rm in rm te N- 154 tid id 30 1 es es 415 429
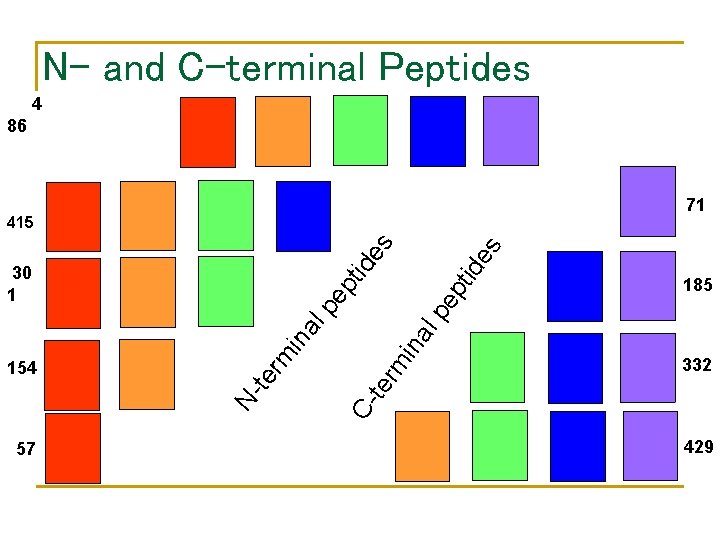
N- and C-terminal Peptides 4 86 71 ep pt 57 185 lp pe ina al 332 C- te rm in rm te N- 154 tid id 30 1 es es 415 429
N- and C-terminal Peptides 4 86 415 71 30 1 185 154 332 57 429

N- and C-terminal Peptides 4 86 71 415 30 1 Reconstruct peptide from the set of masses of fragment ions (mass-spectrum) 185 154 332 57 429
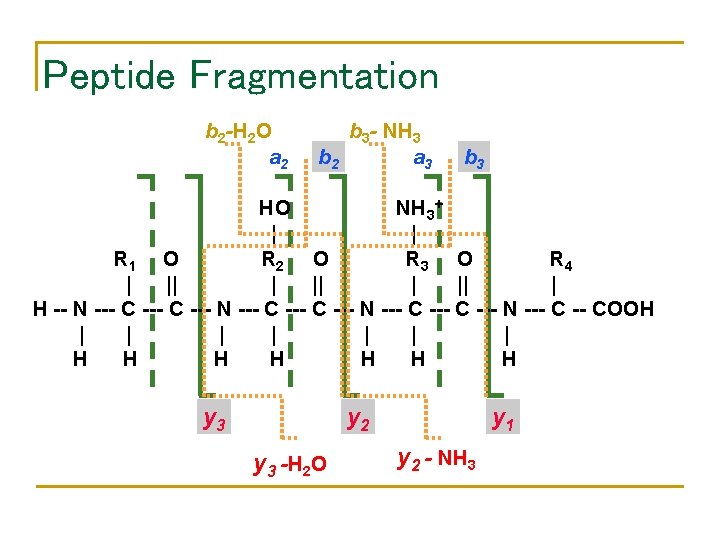
Peptide Fragmentation b 2 -H 2 O a 2 b 3 - NH 3 b 2 a 3 b 3 HO NH 3+ | | R 1 O R 2 O R 3 O R 4 | || | H -- N --- C --- N --- C -- COOH | | | | H H H H y 3 y 2 y 3 -H 2 O y 1 y 2 - NH 3
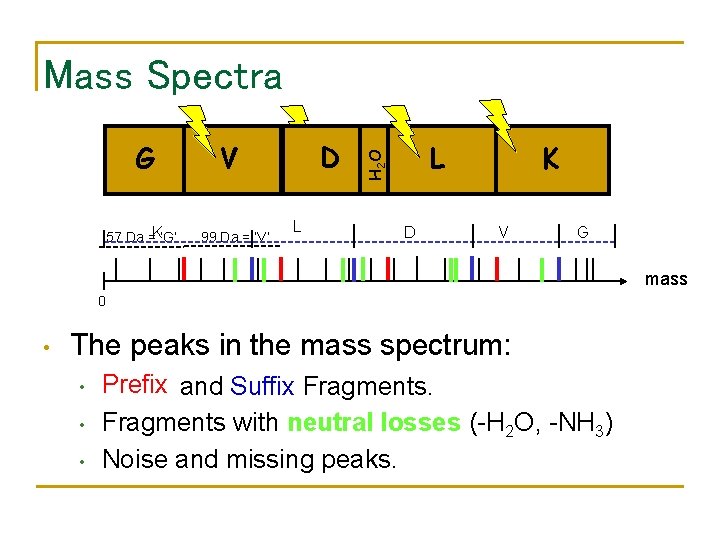
Mass Spectra 57 Da =K‘G’ D D V 99 Da = ‘V’ L L H 2 O G D K V G mass 0 • The peaks in the mass spectrum: • • • Prefix and Suffix Fragments with neutral losses (-H 2 O, -NH 3) Noise and missing peaks.

Protein Identification with MS/MS G V D K Peptide Identification: Intensity MS/MS L mass 00
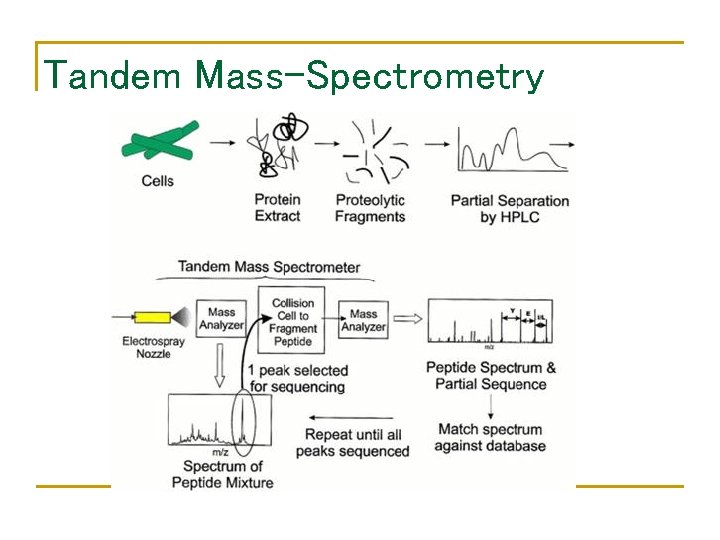
Tandem Mass-Spectrometry
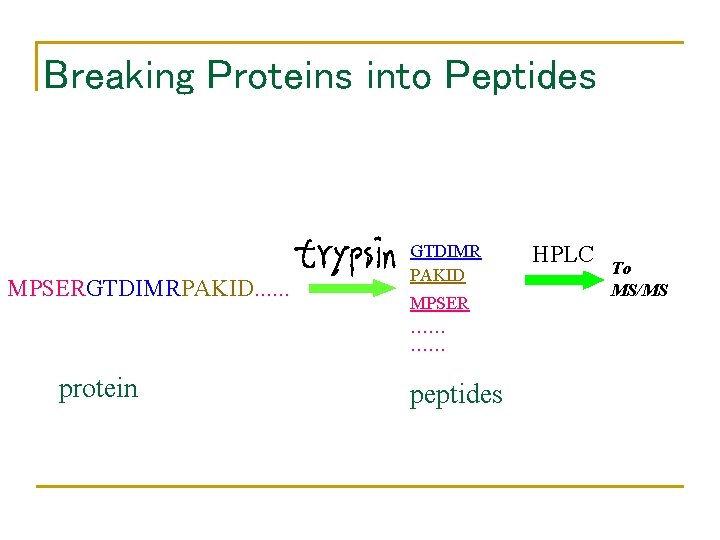
Breaking Proteins into Peptides MPSERGTDIMRPAKID. . . protein GTDIMR PAKID MPSER …… …… peptides HPLC To MS/MS
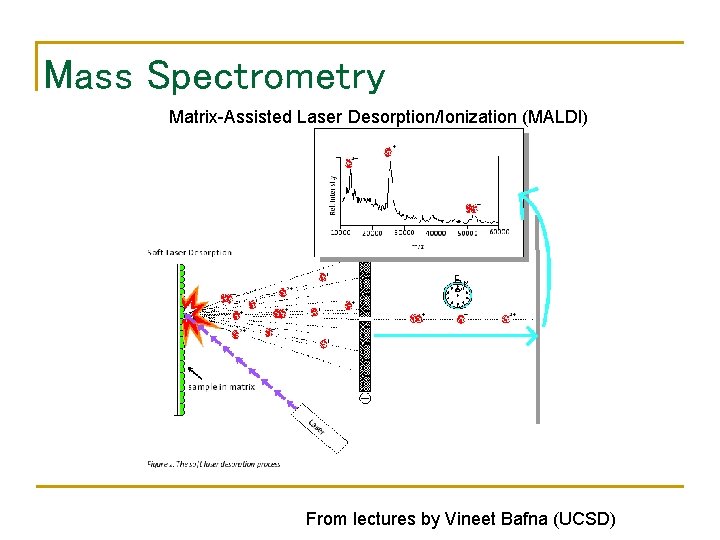
Mass Spectrometry Matrix-Assisted Laser Desorption/Ionization (MALDI) From lectures by Vineet Bafna (UCSD)
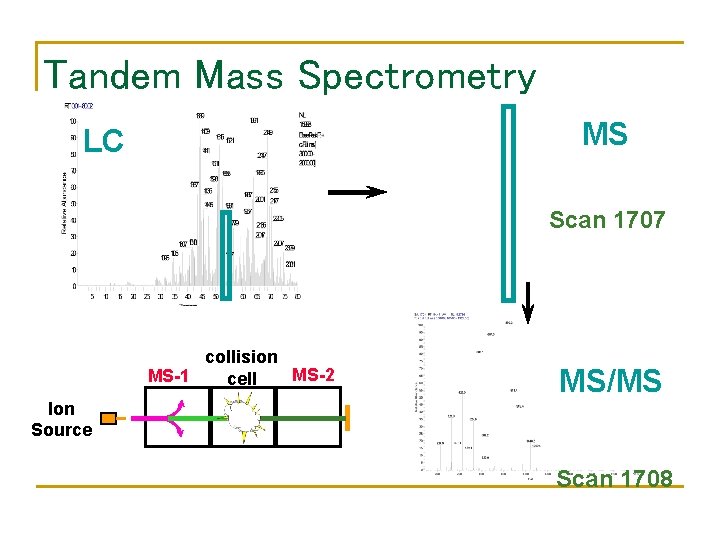
Tandem Mass Spectrometry MS LC Scan 1707 collision MS-2 MS-1 cell MS/MS Ion Source Scan 1708
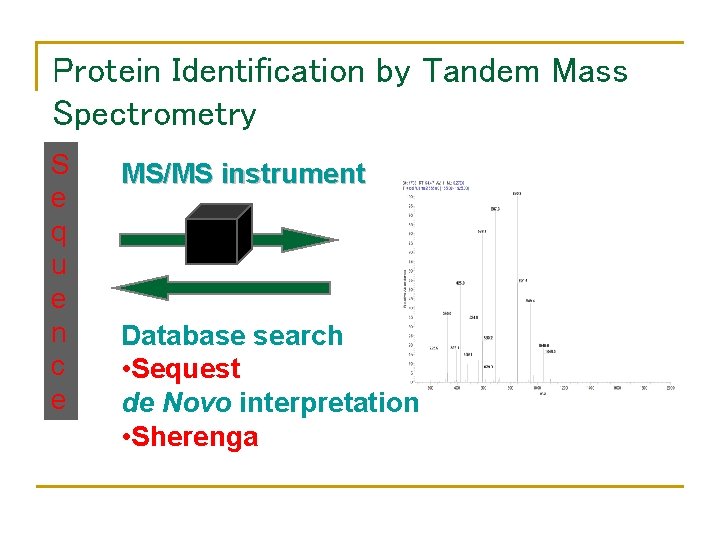
Protein Identification by Tandem Mass Spectrometry S e q u e n c e MS/MS instrument Database search • Sequest de Novo interpretation • Sherenga
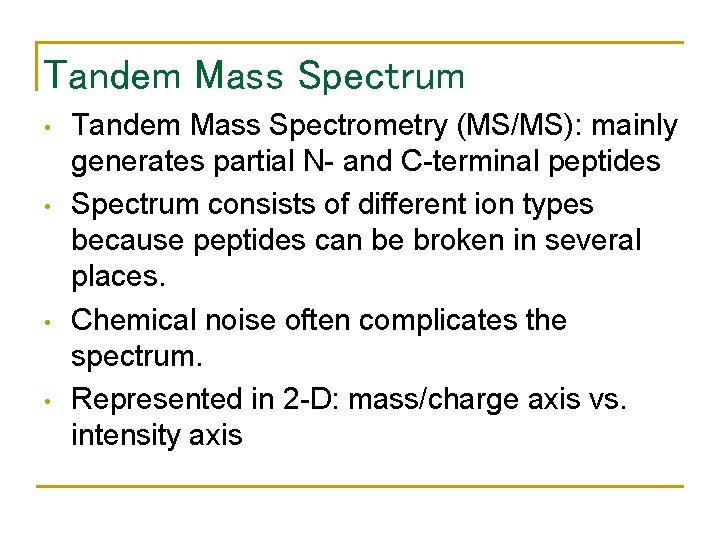
Tandem Mass Spectrum • • Tandem Mass Spectrometry (MS/MS): mainly generates partial N- and C-terminal peptides Spectrum consists of different ion types because peptides can be broken in several places. Chemical noise often complicates the spectrum. Represented in 2 -D: mass/charge axis vs. intensity axis
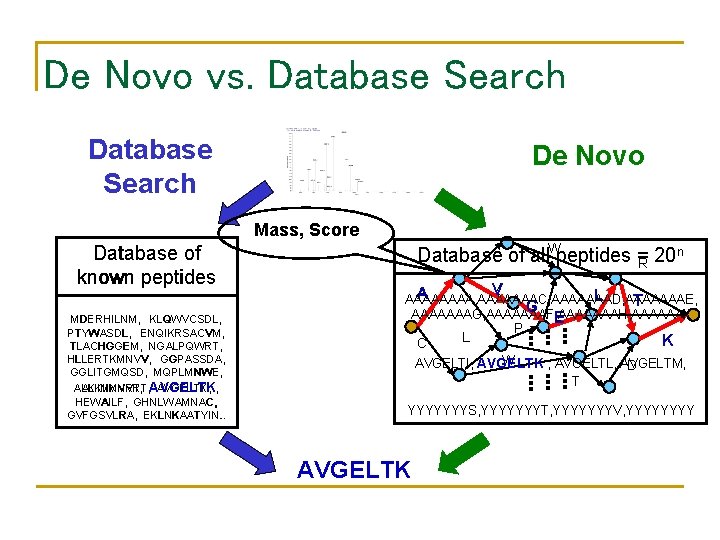
De Novo vs. Database Search De Novo Mass, Score Database of known peptides MDERHILNM, KLQWVCSDL, PTYWASDL, ENQIKRSACVM, TLACHGGEM, NGALPQWRT, HLLERTKMNVV, GGPASSDA, GGLITGMQSD, MQPLMNWE, ALKIIMNVRT, AVGELTK, , HEWAILF, GHNLWAMNAC, GVFGSVLRA, EKLNKAATYIN. . n Database of all. Wpeptides = R 20 V A L AAAA, AAAAAAAC, AAAAAAAD, AAAAAAAE, T G G AAAAAAAG, AAAAAAAF, AAAAAAAH, AAAAAAI, E C L P K K W AVGELTI, AVGELTK , AVGELTL, AVGELTM, D T YYYYYYYS, YYYYYYYT, YYYYYYYV, YYYY AVGELTK
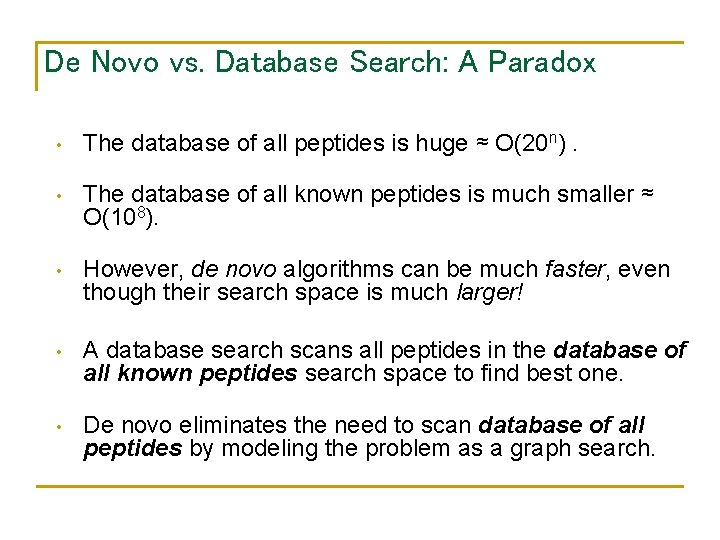
De Novo vs. Database Search: A Paradox • The database of all peptides is huge ≈ O(20 n). • The database of all known peptides is much smaller ≈ O(108). • However, de novo algorithms can be much faster, even though their search space is much larger! • A database search scans all peptides in the database of all known peptides search space to find best one. • De novo eliminates the need to scan database of all peptides by modeling the problem as a graph search.
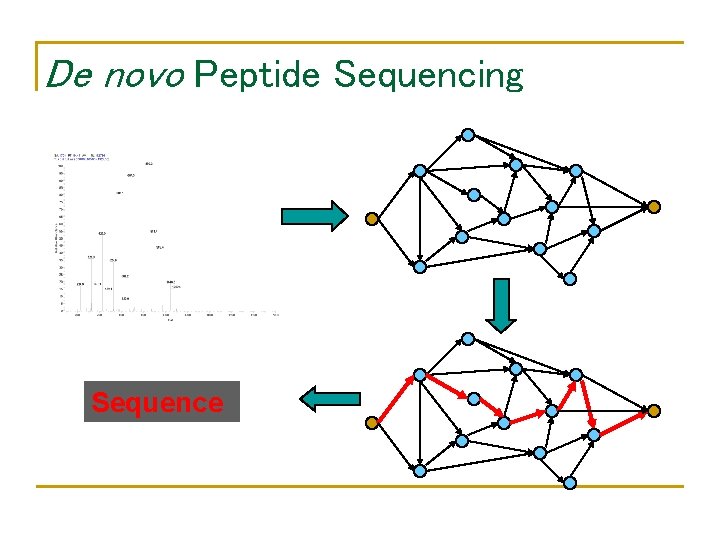
De novo Peptide Sequencing Sequence
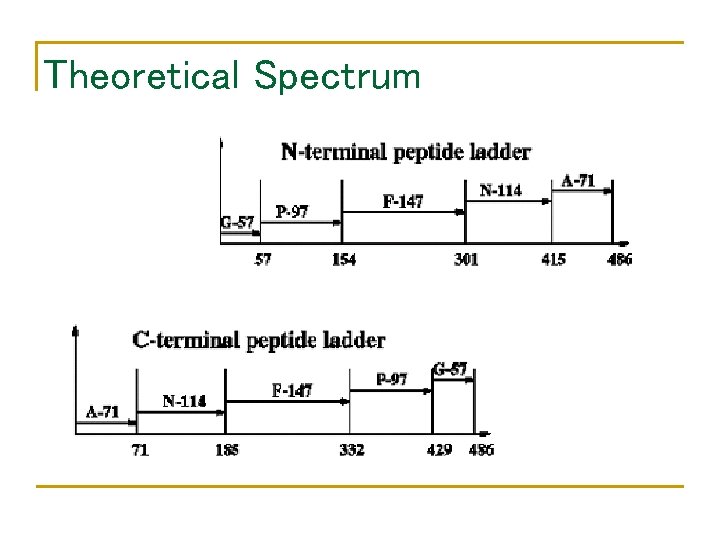
Theoretical Spectrum
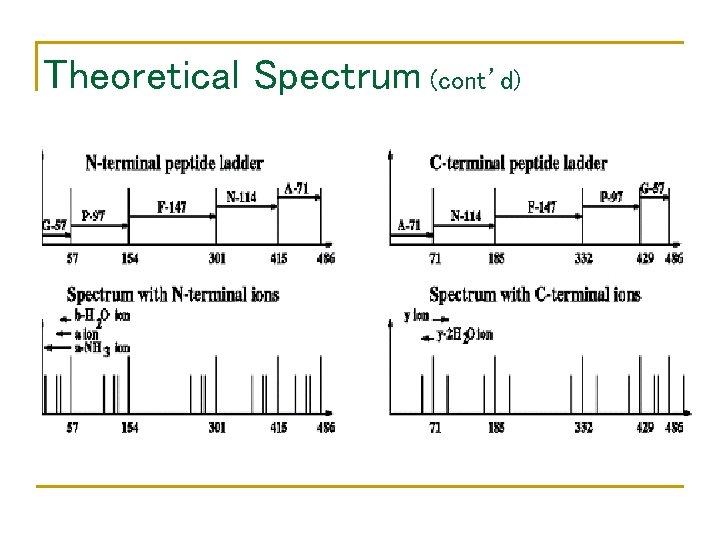
Theoretical Spectrum (cont’d)
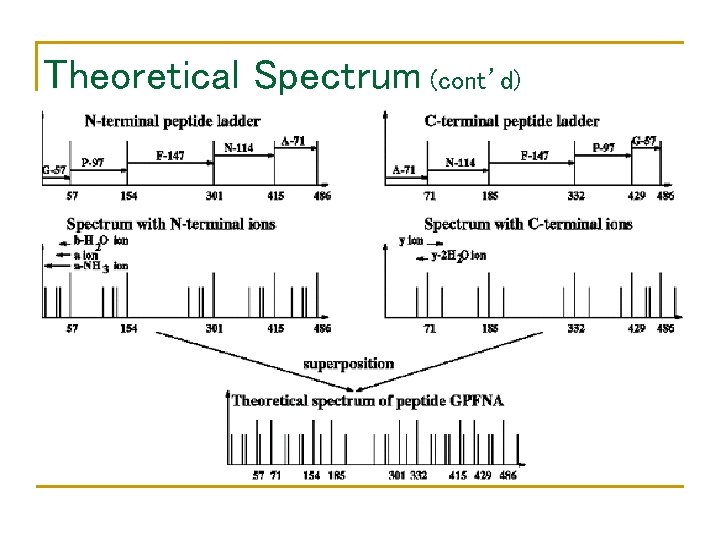
Theoretical Spectrum (cont’d)
Building Spectrum Graph • How to create vertices (from masses) • How to create edges (from mass differences) • How to score paths • How to find best path
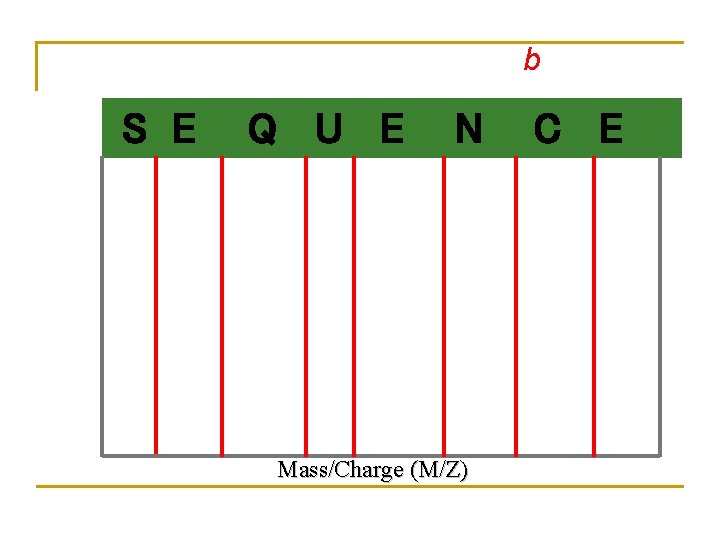
b S E Q U E N Mass/Charge (M/Z) C E
a SE Q U E N Mass/Charge (M/Z) C E
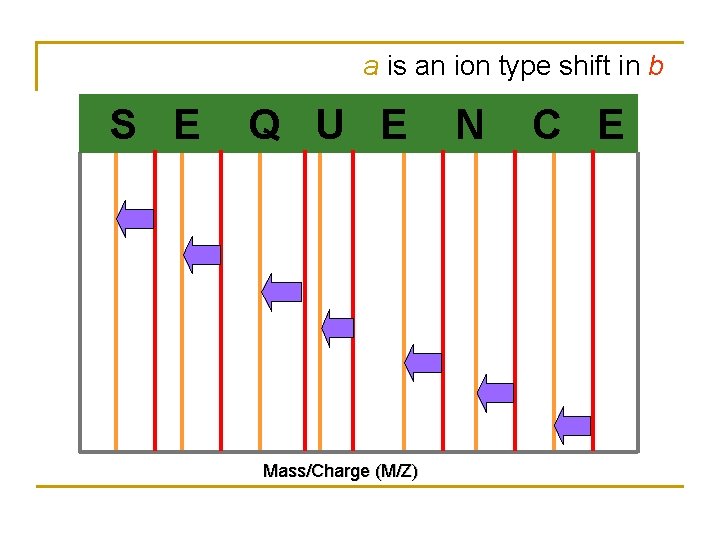
a is an ion type shift in b S E Q U E Mass/Charge (M/Z) N C E
y E C N E U Q Mass/Charge (M/Z) E S
Intensity Mass/Charge (M/Z)
Intensity Mass/Charge (M/Z)

noise Mass/Charge (M/Z)

Intensity MS/MS Spectrum Mass/Charge (M/z)
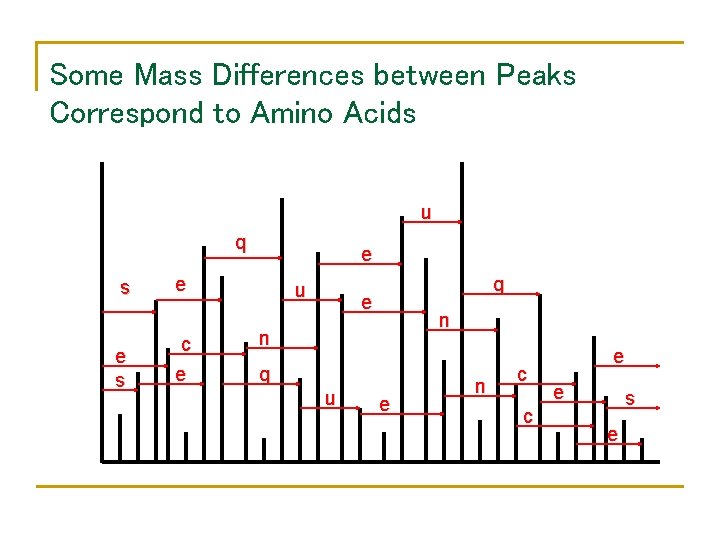
Some Mass Differences between Peaks Correspond to Amino Acids u q s e e c e u q e n n q u e n c c e e s e
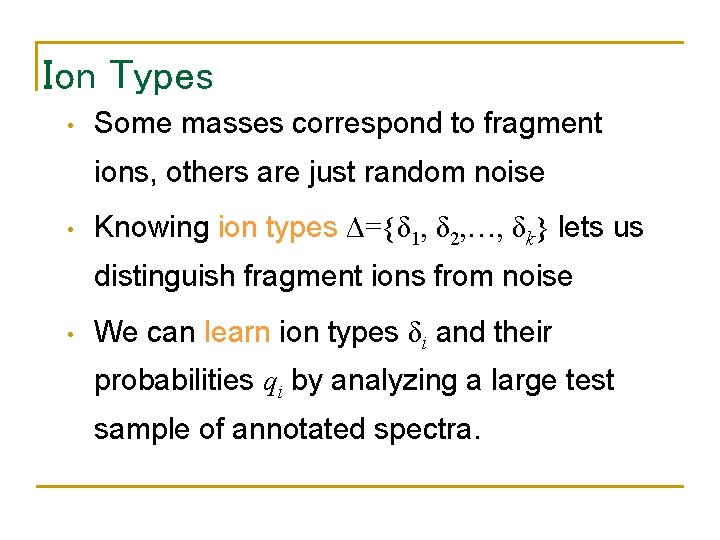
Ion Types • Some masses correspond to fragment ions, others are just random noise • Knowing ion types Δ={δ 1, δ 2, …, δk} lets us distinguish fragment ions from noise • We can learn ion types δi and their probabilities qi by analyzing a large test sample of annotated spectra.
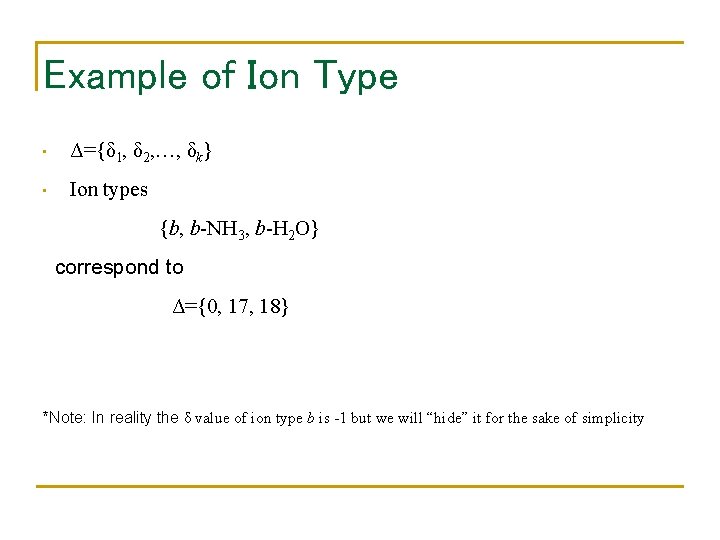
Example of Ion Type • Δ={δ 1, δ 2, …, δk} • Ion types {b, b-NH 3, b-H 2 O} correspond to Δ={0, 17, 18} *Note: In reality the δ value of ion type b is -1 but we will “hide” it for the sake of simplicity
Match between Spectra and the Shared Peak Count • The match between two spectra is the number of masses (peaks) they share (Shared Peak Count or SPC) • In practice mass-spectrometrists use the weighted SPC that reflects intensities of the peaks • Match between experimental and theoretical spectra is defined similarly
Peptide Sequencing Problem Goal: Find a peptide with maximal match between an experimental and theoretical spectrum. Input: • S: experimental spectrum • Δ: set of possible ion types • m: parent mass Output: • P: peptide with mass m, whose theoretical spectrum matches the experimental S spectrum the best
Vertices of Spectrum Graph • Masses of potential N-terminal peptides • Vertices are generated by reverse shifts corresponding to ion types Δ={δ 1, δ 2, …, δk} • Every N-terminal peptide can generate up to k ions m-δ 1, m-δ 2, …, m-δk • Every mass s in an MS/MS spectrum generates k vertices V(s) = {s+δ 1, s+δ 2, …, s+δk} corresponding to potential N-terminal peptides • Vertices of the spectrum graph: {initial vertex} V(s 1) V(s 2) . . . V(sm) {terminal vertex}
Reverse Shifts Shift in H 2 O+NH 3
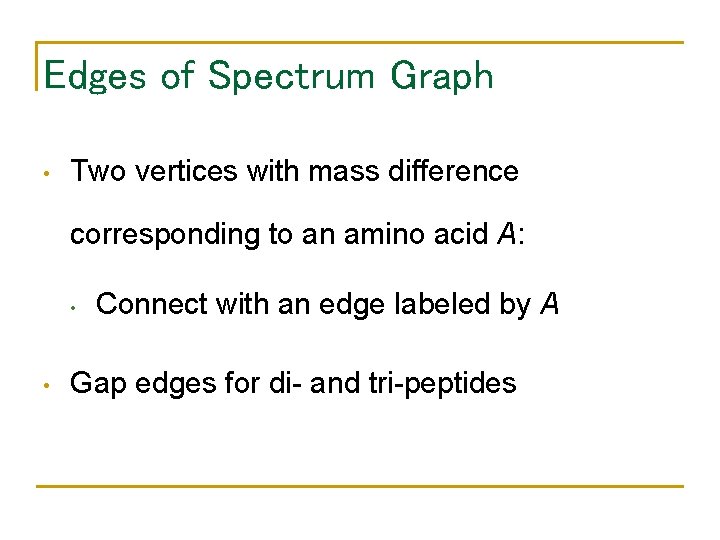
Edges of Spectrum Graph • Two vertices with mass difference corresponding to an amino acid A: • • Connect with an edge labeled by A Gap edges for di- and tri-peptides

Paths • Path in the labeled graph spell out amino acid sequences • There are many paths, how to find the correct one? • We need scoring to evaluate paths
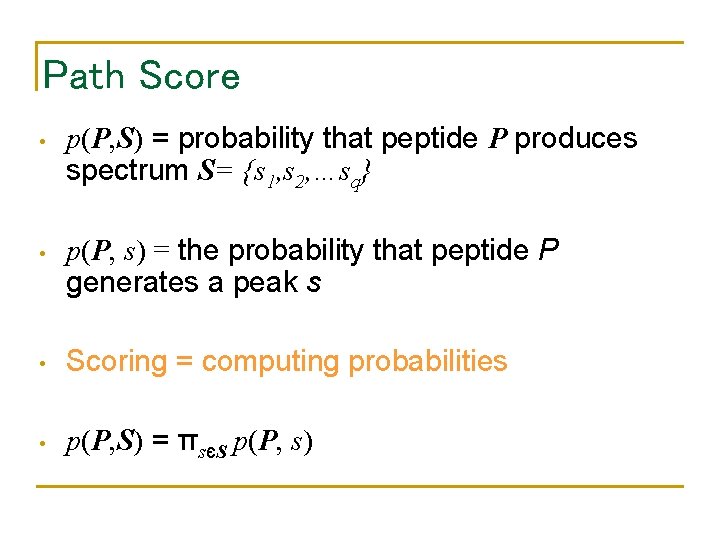
Path Score • p(P, S) = probability that peptide P produces spectrum S= {s 1, s 2, …sq} • p(P, s) = the probability that peptide P generates a peak s • Scoring = computing probabilities • p(P, S) = πsєS p(P, s)
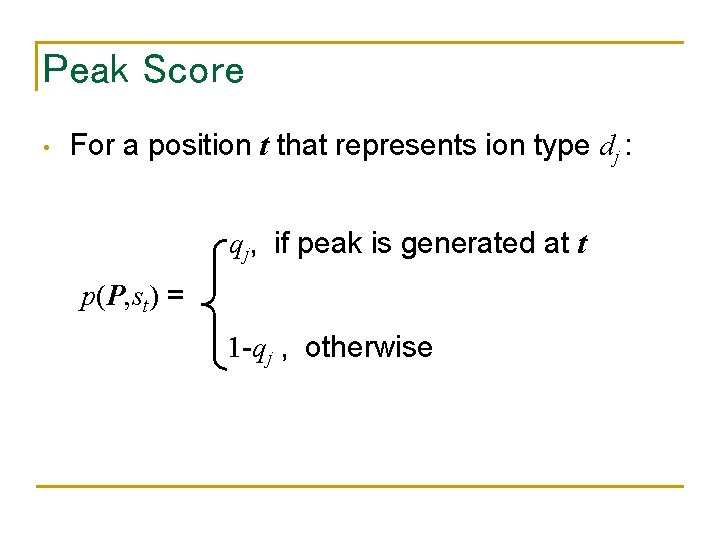
Peak Score • For a position t that represents ion type dj : qj, if peak is generated at t p(P, st) = 1 -qj , otherwise
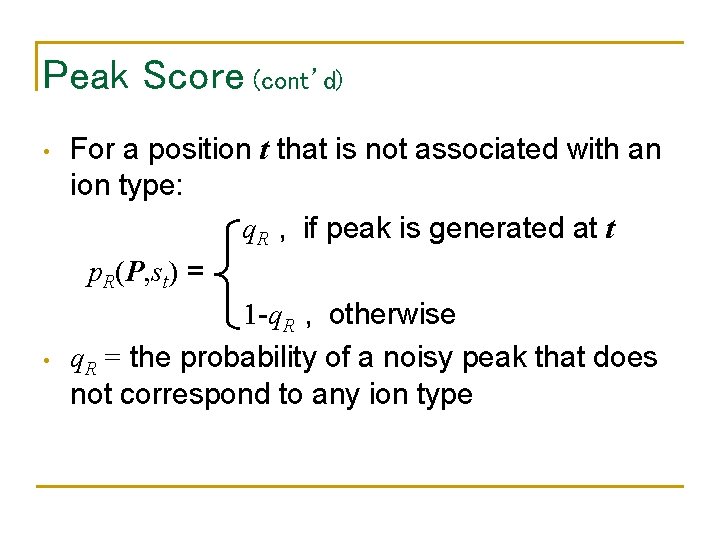
Peak Score (cont’d) • • For a position t that is not associated with an ion type: q. R , if peak is generated at t p. R(P, st) = 1 -q. R , otherwise q. R = the probability of a noisy peak that does not correspond to any ion type
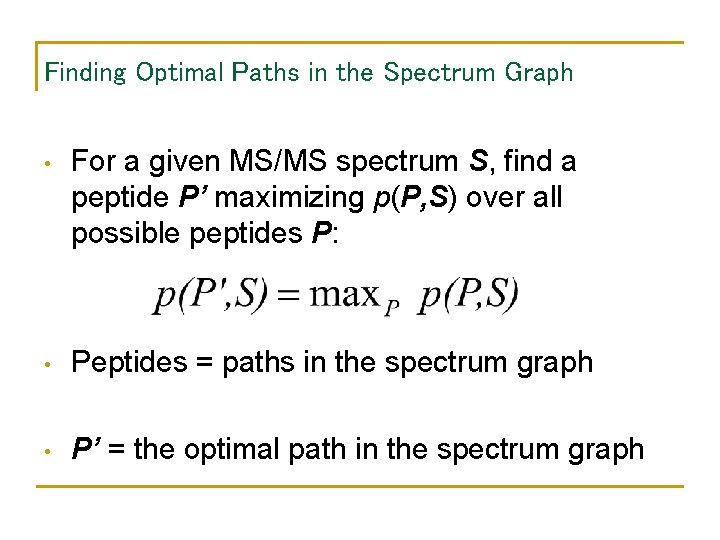
Finding Optimal Paths in the Spectrum Graph • For a given MS/MS spectrum S, find a peptide P’ maximizing p(P, S) over all possible peptides P: • Peptides = paths in the spectrum graph • P’ = the optimal path in the spectrum graph
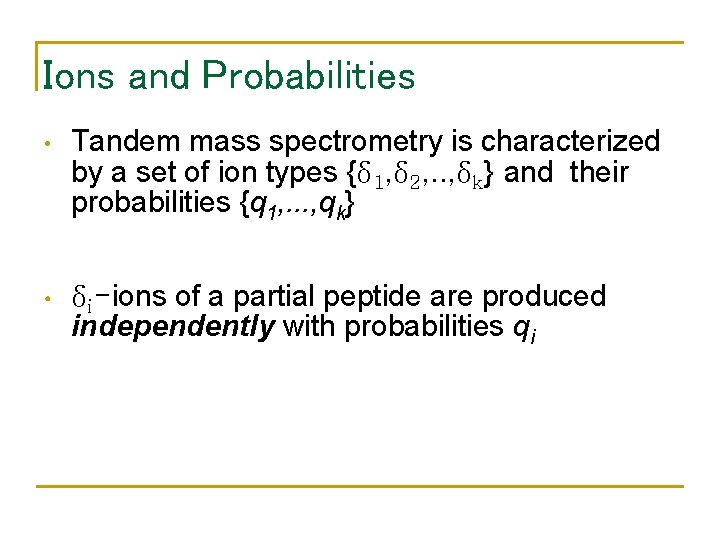
Ions and Probabilities • Tandem mass spectrometry is characterized by a set of ion types {δ 1, δ 2, . . , δk} and their probabilities {q 1, . . . , qk} • δi-ions of a partial peptide are produced independently with probabilities qi
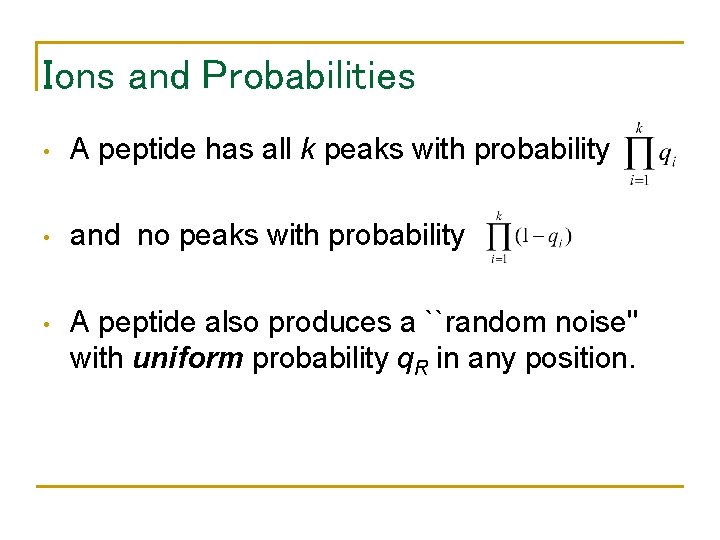
Ions and Probabilities • A peptide has all k peaks with probability • and no peaks with probability • A peptide also produces a ``random noise'' with uniform probability q. R in any position.
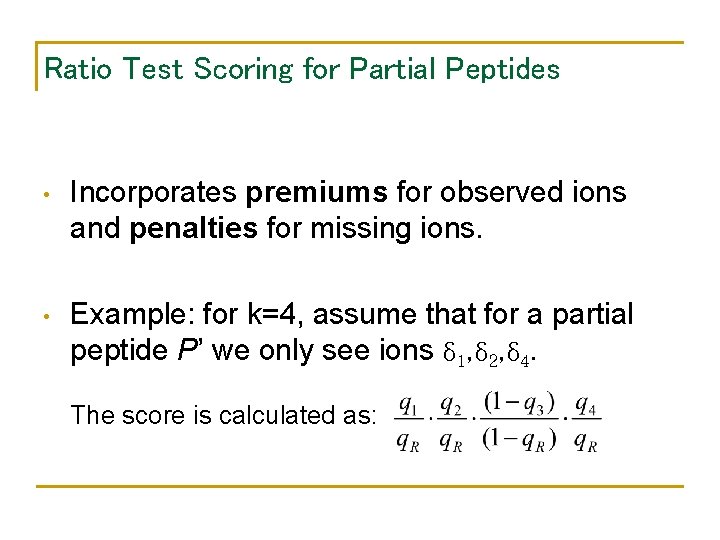
Ratio Test Scoring for Partial Peptides • Incorporates premiums for observed ions and penalties for missing ions. • Example: for k=4, assume that for a partial peptide P’ we only see ions δ 1, δ 2, δ 4. The score is calculated as:
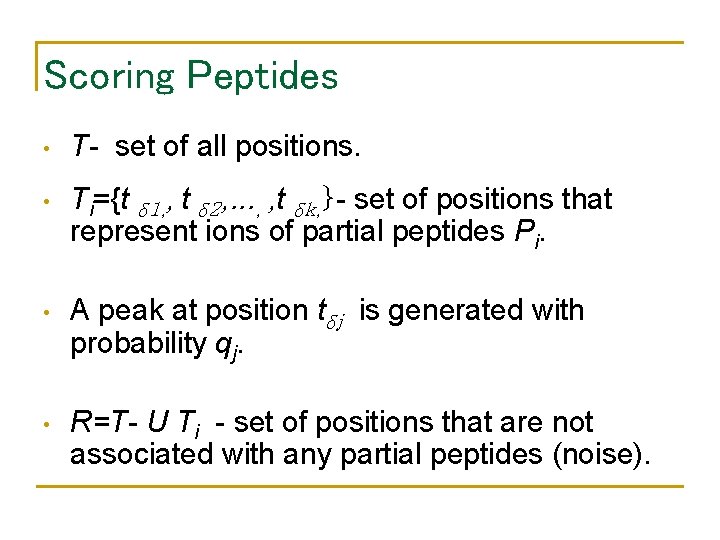
Scoring Peptides • T- set of all positions. • Ti={t δ 1, , t δ 2, . . . , , t δk, }- set of positions that represent ions of partial peptides Pi. • A peak at position tδj is generated with probability qj. • R=T- U Ti - set of positions that are not associated with any partial peptides (noise).
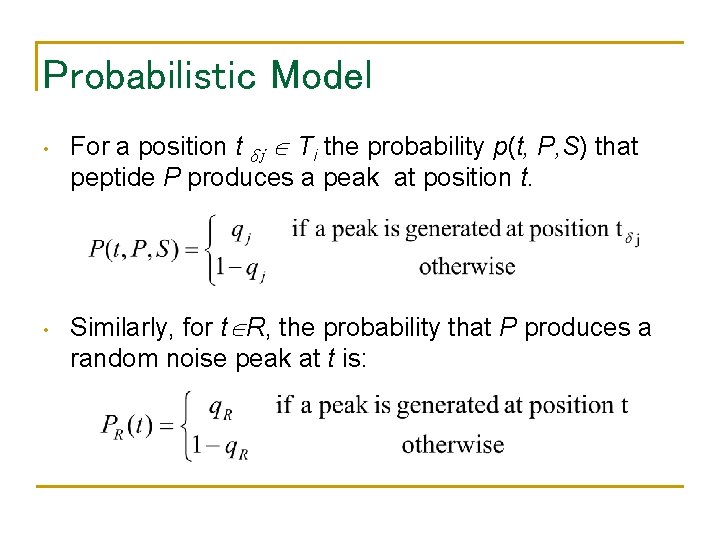
Probabilistic Model • • For a position t δj Ti the probability p(t, P, S) that peptide P produces a peak at position t. Similarly, for t R, the probability that P produces a random noise peak at t is:
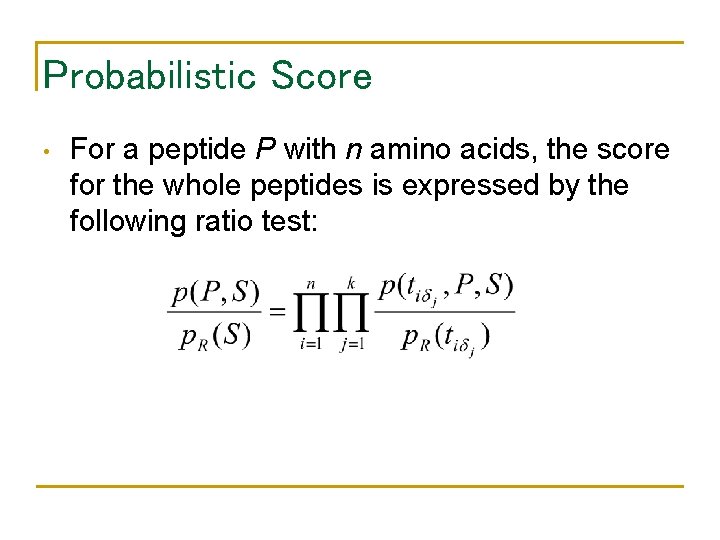
Probabilistic Score • For a peptide P with n amino acids, the score for the whole peptides is expressed by the following ratio test:
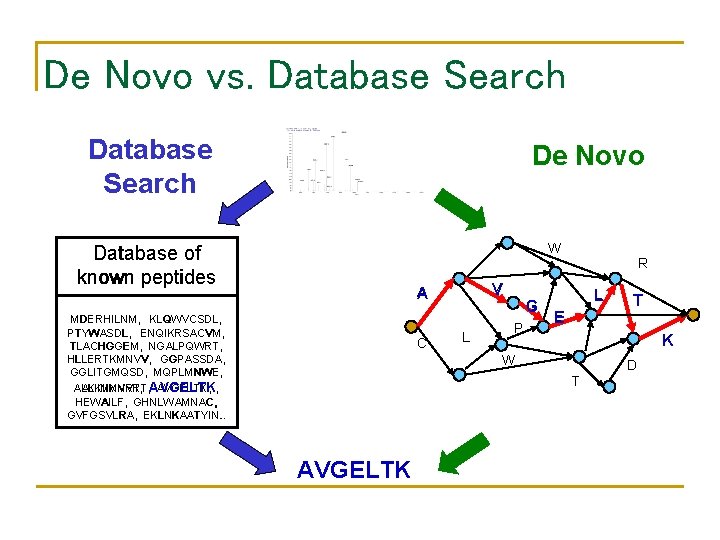
De Novo vs. Database Search De Novo W Database of known peptides V A A MDERHILNM, KLQWVCSDL, PTYWASDL, ENQIKRSACVM, TLACHGGEM, NGALPQWRT, HLLERTKMNVV, GGPASSDA, GGLITGMQSD, MQPLMNWE, ALKIIMNVRT, AVGELTK, , HEWAILF, GHNLWAMNAC, GVFGSVLRA, EKLNKAATYIN. . C L G G P R L L E K K W D T AVGELTK T
- Slides: 55