Protein Sequence and Structurebased Similarity Networks Ioannis Valavanis

Protein Sequence- and Structure-based Similarity Networks Ioannis Valavanis 1, George Spyrou 2, Konstantina Nikita 1 1 Biomedical Simulations and Imaging Laboratory, School of Electrical and Computer Engineering, National Technical University of Athens, Greece 2 Biomedical Research Foundation of the Academy of Athens, Greece Department of Informatics and Communications, University of Athens, Greece Athens, May 7 th, 2008

Outline § Introduction § Construction of Similarity Networks § Analysis of Similarity Networks § Conclusions § Future Work

Networks and Real World Systems Introduction Similarity Networks § Network: Set of vertices (elements of system) and edges (interrelations between elements) § Each real world system can be described by a Network ü ü ü § Quantification of a Network (N (N vertices, K edges) reveals information on the System ü ü Construction § Biological and Chemical Systems Social Interacting Species Computer Networks and Internet Network Degree k=2 K/N Sparsity S = 2 K/(N(N-1)) Centrality measurements of each vertex (degree of a vertex, betweeness of a vertex) Randomness or Regularity based on average minimum path length (L (L) and clustering coefficient (C (C) Most real world systems behave like Small World Networks (SWNs) (Strogatz, 2001) Similarity a) Regular: Great L and C b) Small World Network: Small or intermediate L and great C Networks Analysis Conclusions Future Work c) Random: Small L and C § Few vertices (hubs) dominate the network activity (Barabasi 2003) in SWNs § Some known SWNs: ü ü ü Film Actors Peer to Peer Network Metabolic Network Yeast Protein Interactions Functional Cortical Connectivity Network Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
Network-based description of Proteins Introduction § Folded proteins were transformed to networks (Vendruscolo et al. , 2002; Greene and Higman, 2003) üvertices: residues üedges: given a distance criterion Similarity Networks üProtein Network: SWN Construction § Residue closeness in residue interaction graphs identified functional residues (Amitai et al. , 2004) Similarity Networks Analysis § Similarity proteins networks were constructed and affinity of a protein in the network classified the protein in superfamily, family and fold level (Camoglu et al. , 2006) Conclusions § Structural similarity network of representative proteins of folds was shown to be a SWN Future Work (Sun et al. , 2006) Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008

Current Study Introduction § Quantify the structure of both protein sequence and structural similarity networks for several criteria Similarity Networks Construction § Compare protein sequence and structural similarity network at same sparsity § Find hubs in protein sequence and structural similarity network and their biological significance Similarity Networks Analysis § Extend results to “fold” sequence and structural similarity networks § Assess the information level of sequence-derived features in comparison with Conclusions structures in terms of fold and class assignment Future Work Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
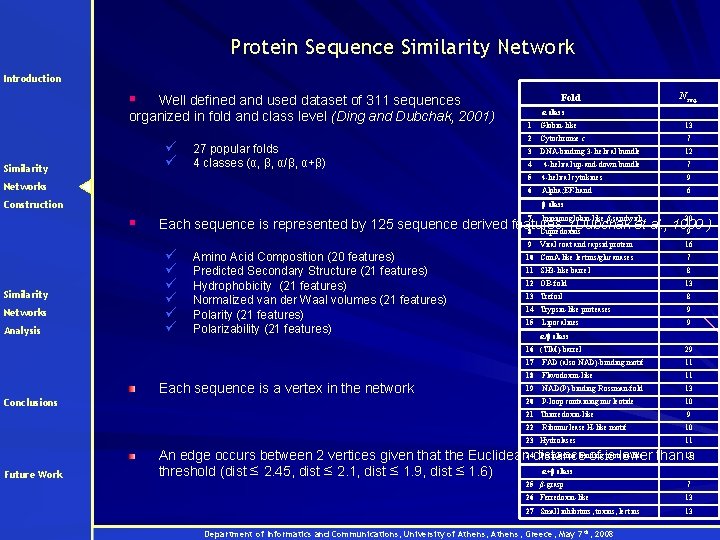
Protein Sequence Similarity Network Introduction § Well defined and used dataset of 311 sequences organized in fold and class level (Ding and Dubchak, 2001) ü ü Similarity 27 popular folds 4 classes (α, β, α/β, α+β) Networks Fold α class 1 Globin-like 2 Cytochrome c 7 3 DNA-binding 3 - helical bundle 12 4 4 -helical up-and-down bundle 7 5 4 -helical cytokines 9 6 Alpha; EF-hand 6 Construction Networks Analysis 7 Immunoglobin-like β-sandwich 30 9 Viral coat and capsid protein 16 10 Con. A-like lectins/glucanases 7 Each sequence is represented by 125 sequence derived features (Dubchak et al. , 1999 ) 8 Cupredoxins 9 ü ü ü Amino Acid Composition (20 features) Predicted Secondary Structure (21 features) Hydrophobicity (21 features) Normalized van der Waal volumes (21 features) Polarity (21 features) Polarizability (21 features) Each sequence is a vertex in the network Conclusions 11 SH 3 -like barrel 8 12 OB-fold 13 13 Trefoil 8 14 Trypsin-like proteases 9 15 9 Lipocalines α/β class 16 (TIM)-barrel 29 17 FAD (also NAD)-binding motif 11 18 Flavodoxin-like 11 19 NAD(P)-binding Rossman-fold 13 20 P-loop containing nucleotide 10 21 Thioredoxin-like 9 22 10 Ribonuclease H-like motif 23 Hydrolases Future Work 13 β class § Similarity Nseq. 11 Periplasmic binding protein-like An edge occurs between 2 vertices given that the Euclidean 24 distance of is lower than 11 a α+β class threshold (dist ≤ 2. 45, dist ≤ 2. 1, dist ≤ 1. 9, dist ≤ 1. 6) 25 β-grasp 7 26 Ferredoxin-like 13 27 Small inhibitors, toxins, lectins 13 Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
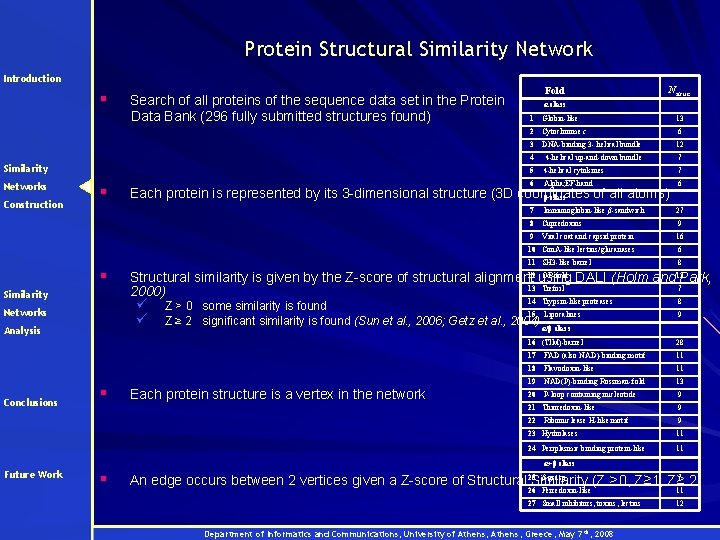
Protein Structural Similarity Network Introduction § Search of all proteins of the sequence data set in the Protein Data Bank (296 fully submitted structures found) Fold Nstruc. α class 1 Globin-like 2 Cytochrome c 6 3 DNA-binding 3 - helical bundle 12 4 4 -helical up-and-down bundle 7 Similarity 5 4 -helical cytokines 7 Networks 6 Alpha; EF-hand 6 7 Immunoglobin-like β-sandwich 27 8 Cupredoxins 9 9 Viral coat and capsid protein 16 10 Con. A-like lectins/glucanases 6 Construction § § Similarity 11 SH 3 -like barrel Analysis § 8 Structural similarity is given by the Z-score of structural alignment using DALI (Holm and Park, 13 Trefoil 7 2000) ü ü Networks Conclusions Each protein is represented by its 3 -dimensional structure (3 D coordinates of all atoms) β class 13 12 OB-fold 13 Z > 0 some similarity is found 15 Lipocalines Z ≥ 2 significant similarity is found (Sun et al. , 2006; Getz et al. , 2004) 14 Trypsin-like proteases 8 16 (TIM)-barrel 28 17 FAD (also NAD)-binding motif 11 18 Flavodoxin-like 11 19 NAD(P)-binding Rossman-fold 13 20 P-loop containing nucleotide 9 9 α/β class Each protein structure is a vertex in the network 21 Thioredoxin-like 22 Ribonuclease H-like motif 9 9 23 Hydrolases 11 24 Periplasmic binding protein-like 11 α+β class Future Work § An edge occurs between 2 vertices given a Z-score of Structural Similarity (Z >0, Z≥ 1, Z≥ 7 2) 25 β-grasp 26 Ferredoxin-like 11 27 Small inhibitors, toxins, lectins 12 Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
Quantification of Similarity Networks Introduction § For each Network (N vertices, K edges) we calculated: ü ü Similarity network degree k=2 K/N Sparsity S=2 K/(N(N-1)) fraction of all finite paths Isolated vertices Networks Construction § For each network, we got its fully connected version by removing isolated vertices § Network parameters were calculated once again Similarity Networks Analysis Conclusions Future Work Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
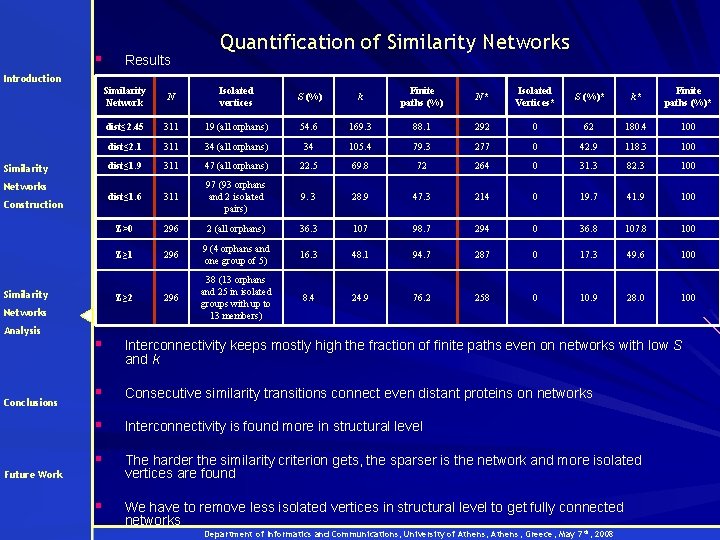
§ Results Quantification of Similarity Networks Introduction Similarity Networks Construction Similarity Conclusions N Isolated vertices S (%) k Finite paths (%) N* Isolated Vertices* S (%)* k* Finite paths (%)* dist≤ 2. 45 311 19 (all orphans) 54. 6 169. 3 88. 1 292 0 62 180. 4 100 dist≤ 2. 1 311 34 (all orphans) 34 105. 4 79. 3 277 0 42. 9 118. 3 100 dist≤ 1. 9 311 47 (all orphans) 22. 5 69. 8 72 264 0 31. 3 82. 3 100 dist≤ 1. 6 311 97 (93 orphans and 2 isolated pairs) 9. 3 28. 9 47. 3 214 0 19. 7 41. 9 100 Z>0 296 2 (all orphans) 36. 3 107 98. 7 294 0 36. 8 107. 8 100 Z≥ 1 296 9 (4 οrphans and one group of 5) 16. 3 48. 1 94. 7 287 0 17. 3 49. 6 100 296 38 (13 orphans and 25 in isolated groups with up to 13 members) 8. 4 24. 9 76. 2 258 0 10. 9 28. 0 100 Z≥ 2 Networks Analysis Similarity Network § Interconnectivity keeps mostly high the fraction of finite paths even on networks with low S and k § Consecutive similarity transitions connect even distant proteins on networks § Interconnectivity is found more in structural level § The harder the similarity criterion gets, the sparser is the network and more isolated vertices are found § We have to remove less isolated vertices in structural level to get fully connected networks Future Work Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
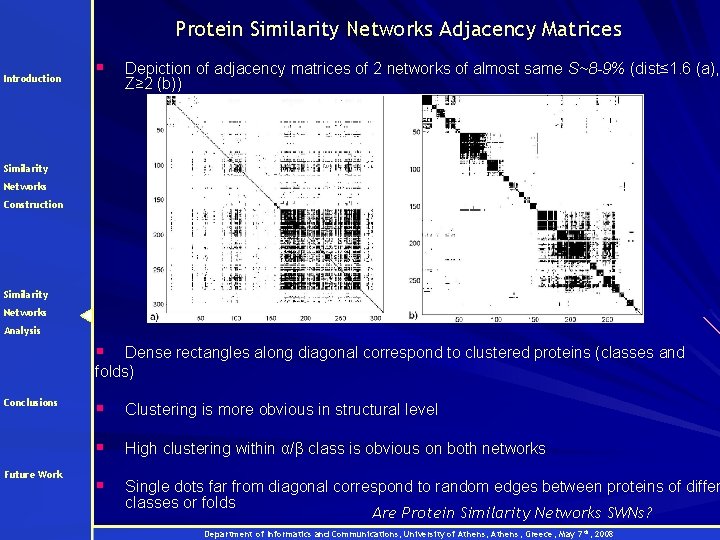
Protein Similarity Networks Adjacency Matrices Introduction § Depiction of adjacency matrices of 2 networks of almost same S~8 -9% (dist≤ 1. 6 (a), Z≥ 2 (b)) Similarity Networks Construction Similarity Networks Analysis § Dense rectangles along diagonal correspond to clustered proteins (classes and folds) Conclusions Future Work § Clustering is more obvious in structural level § High clustering within α/β class is obvious on both networks § Single dots far from diagonal correspond to random edges between proteins of differ classes or folds Are Protein Similarity Networks SWNs? Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
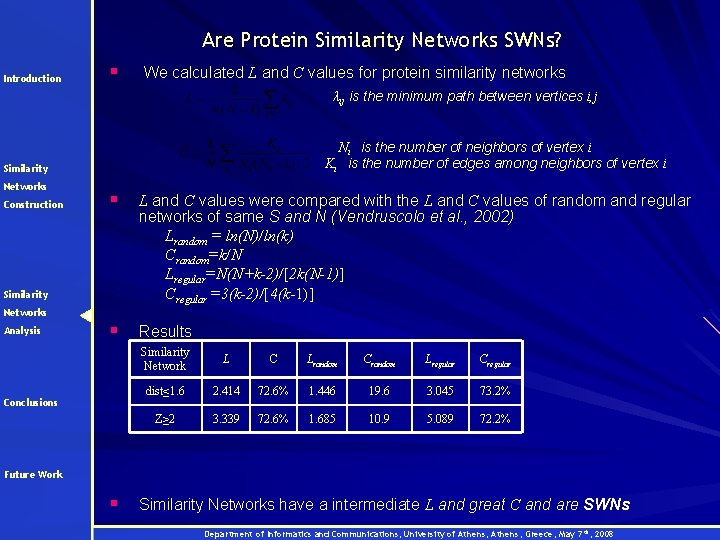
Are Protein Similarity Networks SWNs? Introduction § We calculated L and C values for protein similarity networks λij is the minimum path between vertices i, j Ni is the number of neighbors of vertex i Ki is the number of edges among neighbors of vertex i Similarity Networks Construction § L and C values were compared with the L and C values of random and regular networks of same S and N (Vendruscolo et al. , 2002) Lrandom = ln(N)/ln(k) Crandom=k/N Lregular=N(N+k-2)/[2 k(N-1)] Cregular =3(k-2)/[4(k-1)] § Results Similarity Networks Analysis Conclusions Similarity Network L C Lrandom Crandom Lregular Cregular dist≤ 1. 6 2. 414 72. 6% 1. 446 19. 6 3. 045 73. 2% Z≥ 2 3. 339 72. 6% 1. 685 10. 9 5. 089 72. 2% Future Work § Similarity Networks have a intermediate L and great C and are SWNs Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
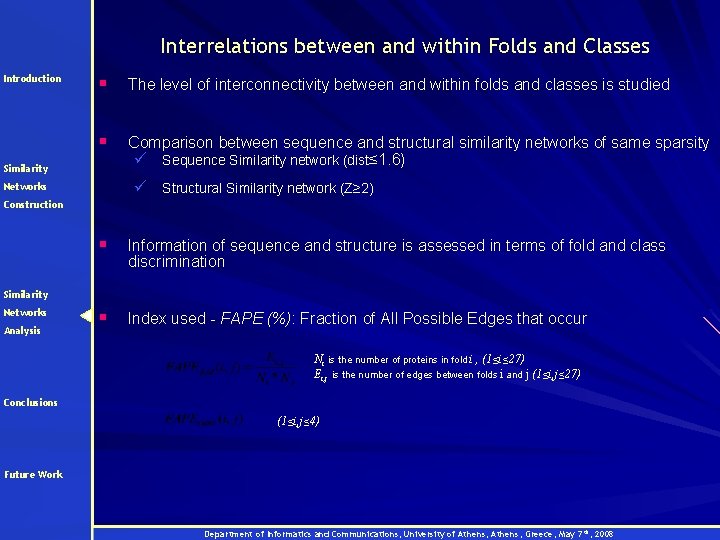
Interrelations between and within Folds and Classes Introduction § The level of interconnectivity between and within folds and classes is studied § Comparison between sequence and structural similarity networks of same sparsity ü Sequence Similarity network (dist≤ 1. 6) Similarity ü Structural Similarity network (Z≥ 2) Networks Construction § Information of sequence and structure is assessed in terms of fold and class discrimination § Index used - FAPE (%): Fraction of All Possible Edges that occur Similarity Networks Analysis Ni is the number of proteins in fold i , (1≤i≤ 27) Ei, j is the number of edges between folds i and j (1≤i, j≤ 27) Conclusions (1≤i, j≤ 4) Future Work Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
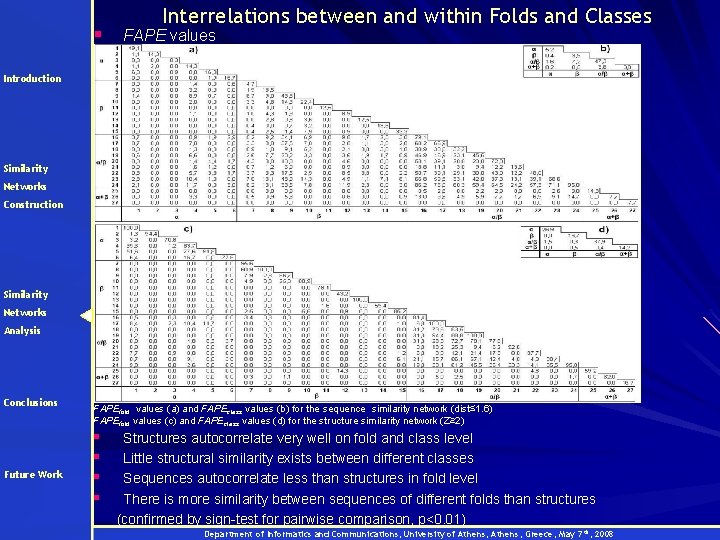
§ Interrelations between and within Folds and Classes FAPE values Introduction Similarity Networks Construction Similarity Networks Analysis Conclusions Future Work FAPEfold values (a) and FAPEclass values (b) for the sequence similarity network (dist≤ 1. 6) FAPEfold values (c) and FAPEclass values (d) for the structure similarity network (Z≥ 2) § § Structures autocorrelate very well on fold and class level Little structural similarity exists between different classes Sequences autocorrelate less than structures in fold level There is more similarity between sequences of different folds than structures (confirmed by sign-test for pairwise comparison, p<0. 01) Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
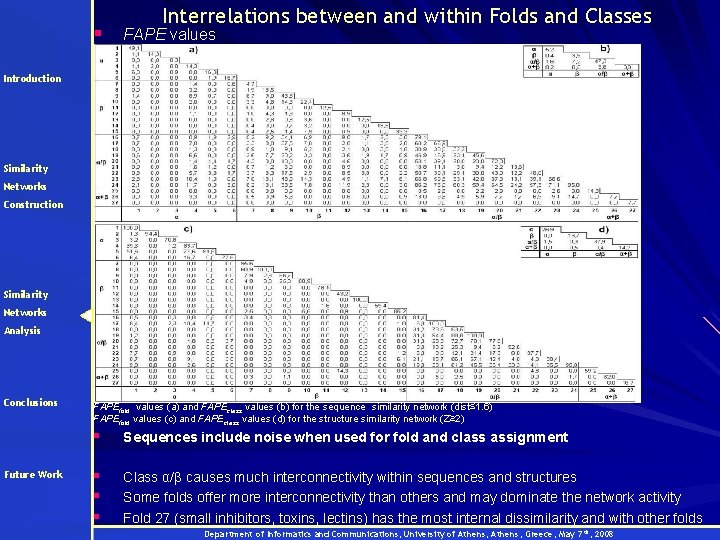
§ Interrelations between and within Folds and Classes FAPE values Introduction Similarity Networks Construction Similarity Networks Analysis Conclusions Future Work FAPEfold values (a) and FAPEclass values (b) for the sequence similarity network (dist≤ 1. 6) FAPEfold values (c) and FAPEclass values (d) for the structure similarity network (Z≥ 2) § Sequences include noise when used for fold and class assignment § § § Class α/β causes much interconnectivity within sequences and structures Some folds offer more interconnectivity than others and may dominate the network activity Fold 27 (small inhibitors, toxins, lectins) has the most internal dissimilarity and with other folds Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
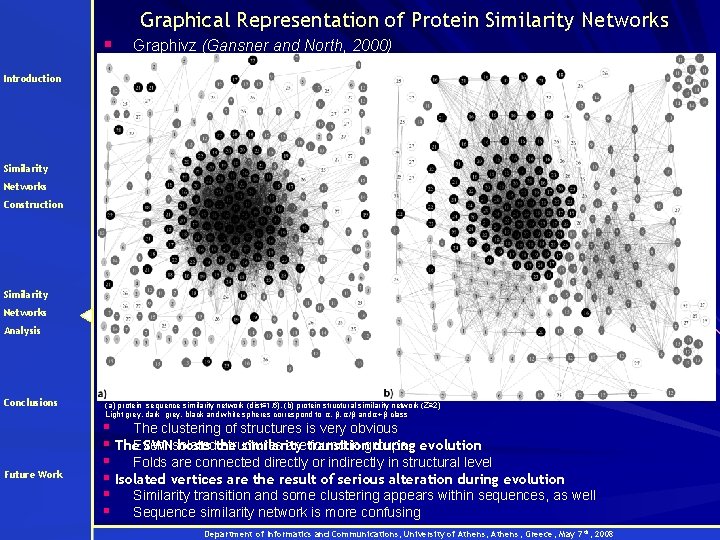
Graphical Representation of Protein Similarity Networks § Graphivz (Gansner and North, 2000) Introduction Similarity Networks Construction Similarity Networks Analysis Conclusions Future Work (a) protein sequence similarity network (dist≤ 1. 6), (b) protein structural similarity network (Z≥ 2) Light grey, dark grey, black and white spheres correspond to α, β, α/β and α+β class § The clustering of structures is very obvious SWNisolated hosts the similarity during evolution § The. Even structures aretransition found in groups § Folds are connected directly or indirectly in structural level § Isolated vertices are the result of serious alteration during evolution § Similarity transition and some clustering appears within sequences, as well § Sequence similarity network is more confusing Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
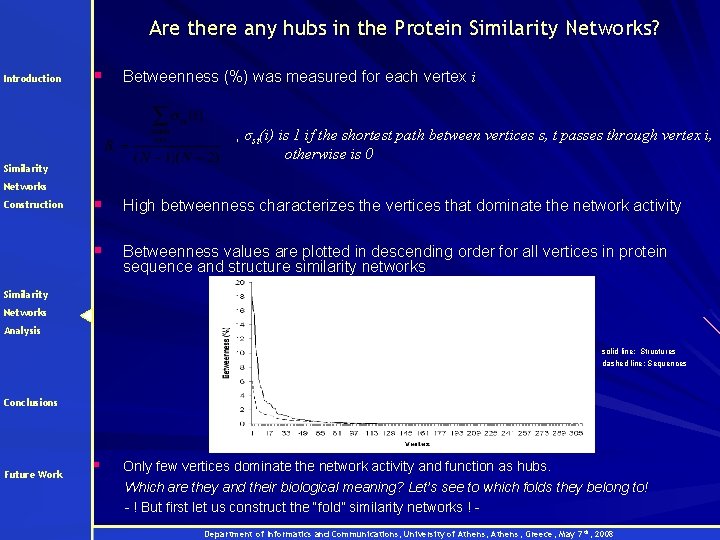
Are there any hubs in the Protein Similarity Networks? Introduction § Betweenness (%) was measured for each vertex i , σst(i) is 1 if the shortest path between vertices s, t passes through vertex i, otherwise is 0 Similarity Networks Construction § High betweenness characterizes the vertices that dominate the network activity § Betweenness values are plotted in descending order for all vertices in protein sequence and structure similarity networks Similarity Networks Analysis solid line: Structures dashed line: Sequences Conclusions Future Work § Only few vertices dominate the network activity and function as hubs. Which are they and their biological meaning? Let’s see to which folds they belong to! - ! But first let us construct the “fold” similarity networks ! Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
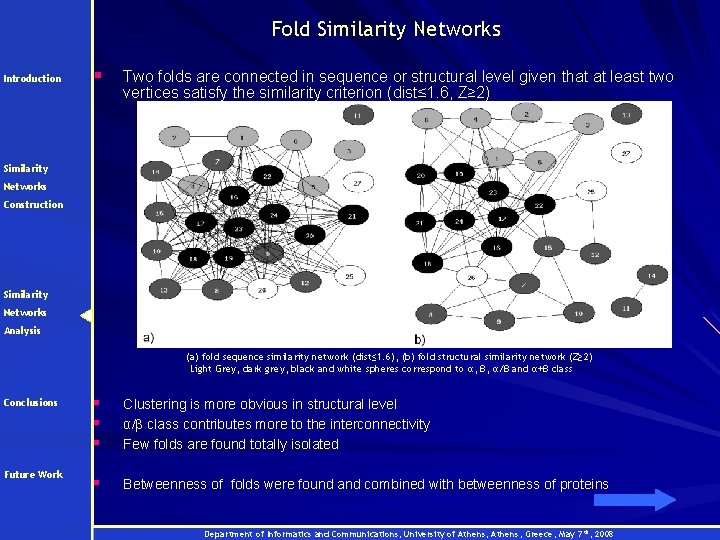
Fold Similarity Networks Introduction § Two folds are connected in sequence or structural level given that at least two vertices satisfy the similarity criterion (dist≤ 1. 6, Z≥ 2) Similarity Networks Construction Similarity Networks Analysis (a) fold sequence similarity network (dist≤ 1. 6), (b) fold structural similarity network (Z≥ 2) Light Grey, dark grey, black and white spheres correspond to α, β, α/β and α+β class Conclusions § § § Clustering is more obvious in structural level α/β class contributes more to the interconnectivity Few folds are found totally isolated Future Work § Betweenness of folds were found and combined with betweenness of proteins Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
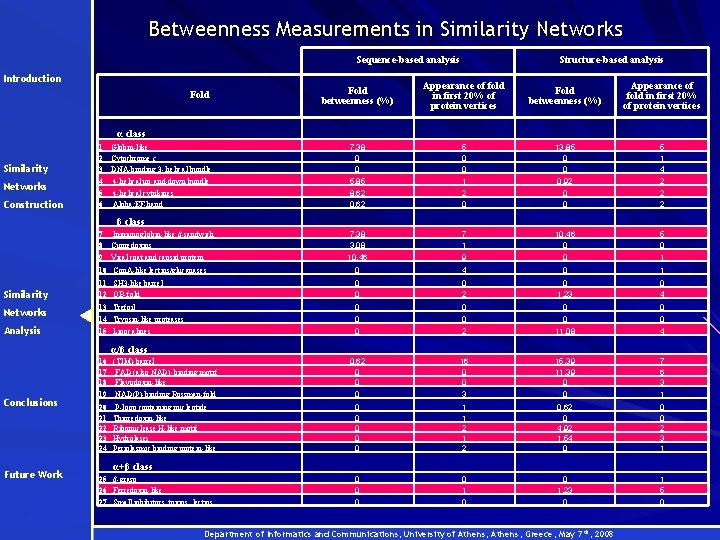
Betweenness Measurements in Similarity Networks Sequence-based analysis Introduction Structure-based analysis Fold betweenness (%) Appearance of fold in first 20% of protein vertices 7. 38 0 0 5. 85 8. 62 0. 62 5 0 0 1 2 0 13. 85 0 0 0. 92 0 0 5 1 4 2 2 2 7. 38 3. 08 10. 46 7 1 9 10. 46 0 0 5 0 1 10 Con. A-like lectins/glucanases 0 4 0 1 11 SH 3 -like barrel 12 OB-fold 0 0 0 2 0 1. 23 0 4 Networks 13 Trefoil 14 Trypsin-like proteases 0 0 0 0 Analysis 15 Lipocalines 0 2 11. 08 4 0. 62 0 0 0 16 0 0 3 15. 39 11. 39 0 0 7 6 3 1 0 0 0 1 1 2 0. 62 0 4. 92 1. 54 0 0 0 2 3 1 0 0 1. 23 0 1 5 0 Fold α class Similarity Networks Construction 1 2 3 4 5 6 Globin-like Cytochrome c DNA-binding 3 - helical bundle 4 -helical up-and-down bundle 4 -helical cytokines Alpha; EF-hand 7 8 9 Immunoglobin-like β-sandwich Cupredoxins Viral coat and capsid protein β class Similarity α/β class Conclusions Future Work 16 (TIM)-barrel 17 FAD (also NAD)-binding motif 18 Flavodoxin-like 19 NAD(P)-binding Rossman-fold 20 21 22 23 24 P-loop containing nucleotide Thioredoxin-like Ribonuclease H-like motif Hydrolases Periplasmic binding protein-like α+β class 25 β-grasp 26 Ferredoxin-like 27 Small inhibitors, toxins, lectins Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
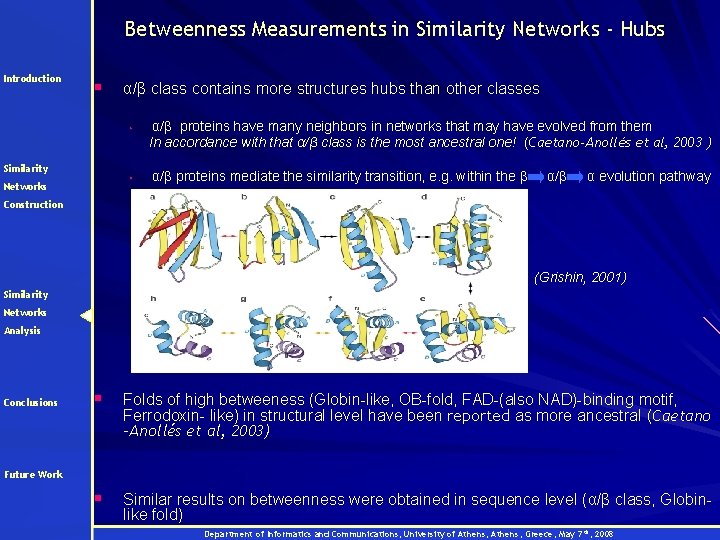
Betweenness Measurements in Similarity Networks - Hubs Introduction § α/β class contains more structures hubs than other classes • Similarity • Networks α/β proteins have many neighbors in networks that may have evolved from them In accordance with that α/β class is the most ancestral one! (Caetano-Anollés et al, 2003 ) α/β proteins mediate the similarity transition, e. g. within the β α/β α evolution pathway Construction (Grishin, 2001) Similarity Networks Analysis Conclusions § Folds of high betweeness (Globin-like, OB-fold, FAD-(also NAD)-binding motif, Ferrodoxin- like) in structural level have been reported as more ancestral (Caetano -Anollés et al, 2003) § Similar results on betweenness were obtained in sequence level (α/β class, Globinlike fold) Future Work Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008

Conclusions Introduction § High interconnectivity was found in both sequence and structural similarity networks § Similarity transition and interconnectivity is more obvious in structural level and Similarity Networks Construction appears due to evolution § Both networks were classified as SWNs, like other real-world systems ü Hubs were found and related to the ancestrality of proteins/folds § Comparison of protein sequence and structure similarity networks at same Similarity Networks Analysis sparsity showed commonalities: üInterconnectivity due to α/β class and certain folds üClustering in folds and classes üIsolated folds on both levels and differences: üMore clustering in structural level üMore interrelation of different folds and classes in sequence level Conclusions The source of noise in sequences when used for fold and class assignment is obvious Future Work § The information quality of sequence derived features is still to be studied and optimized Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
Future Work Introduction § Assess the quality of each of subsets of sequence-derived features used here, e. g. using graph similarity metrics between the two networks Similarity Networks Construction § Extend the pool of used sequence-derived features and optimize the features to use on a global network basis Similarity Networks § Build as similar as possible protein similarity networks on sequence and structure Analysis level Conclusions § Proceed to fold and class assignment using the sequence information in the optimized sequence similarity network, e. g. using graph-based classifiers Future Work Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008

Literature Vendruscolo, M. , Dokholyan, N. V. Paci, E. and Karplus, M. (2002) Small-world view of the amino acids that play a key role in protein folding. Phys. Rev. E. 65, id. : 061910. Introduction Barabasi AL. Linked: How everything is connected to everything else and what it means. New York: Plume Books; 2003. Greene, L. H. and Higman, V. A. (2003) Uncovering Network Systems Within Protein Structures. J. Mol. Biol. , 334, 781791. Similarity Networks Construction Amitai, G. et al. (2004) Network Analysis of Protein Structures Identifies Functional Residues. J. Mol. Biol. , 344, 11351144. Camoglu, O. , Can, T. , Singh, A. K. (2006) Integrating multi-attribute similarity networks for robust representation of the protein space. Bioinformatics, 22, 1585 - 1592. Sun, Z. B. , Zou, X. W. , Guan, W. and Jin, Z. Z. (2006) The architectonic fold similarity network in protein fold space. Eur. Phys. J. B, 49, 127 -134. Ding, C. H. Q and Dubchak, I. (2001) Multi-class protein fold recognition using support vector machines and neural networks. Bioinformatics, 7, 349– 358. Similarity Networks Analysis Dubchak et al. (1999) Recognition of a protein fold in the context of the Structural Classification of Proteins (SCOP) classification. Proteins, 35, 401 -407. Holm, L. and Sander, C. (1994) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. , 233, 123138 Strogatz, S. H. (2001) Exploring complex networks. Nature, 410, 268– 276 Conclusions Future Work Gansner, E. and North, S. (2000) An open graph visualization system and its applications to software engineering. Softw, Pract. Exper. , 30, 1203 -1233. Getz, G. , Starovolsky, A. and Domany, E. (2004) F 2 CS: FSSP to CATH and SCOP prediction server. Bioinformatics, 20, 2150 -2152. Caetano-Anollés G. , Caetano-Anollés D. An evolutionarily structured universe of protein architecture. Genome Res 2003; 13: 1563 -1571. Grishin NV. Fold change in evolution of protein structures. J Struct Biol 2001; 134: 67 -185. Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008

THANK YOU!!!! Introduction QUESTIONS? COMMENTS? Similarity Networks Construction (ivalavan@biosim. ntua. gr) ______________________ Similarity Visit Networks Analysis Conclusions Future Work www. bibe 2008. org for details on 8 th IEEE International Conference on Bio. Informatics and Bio. Engineering (Athens, 8 - 10 October 2008) (Paper Submission deadline: 15/6/2008) Department of Informatics and Communications, University of Athens, Greece, May 7 th, 2008
- Slides: 23