Protein Secondary Structure Bioinformatics Tools and Multiple Sequence
Protein Secondary Structure, Bioinformatics Tools, and Multiple Sequence Alignments Finding Similar Sequences Predicting Secondary Structures Predicting Protein Folds (Tertiary Structures)

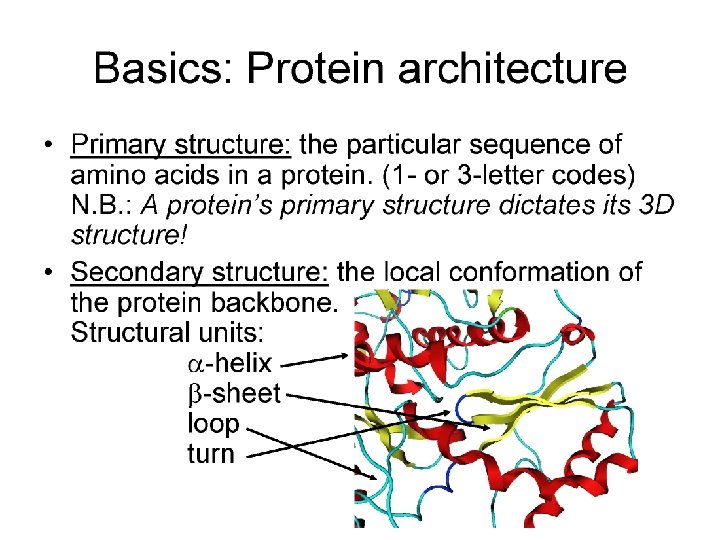
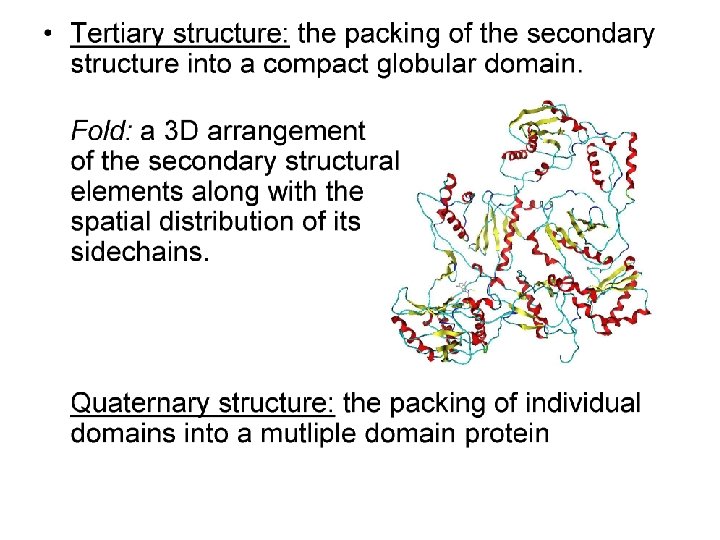
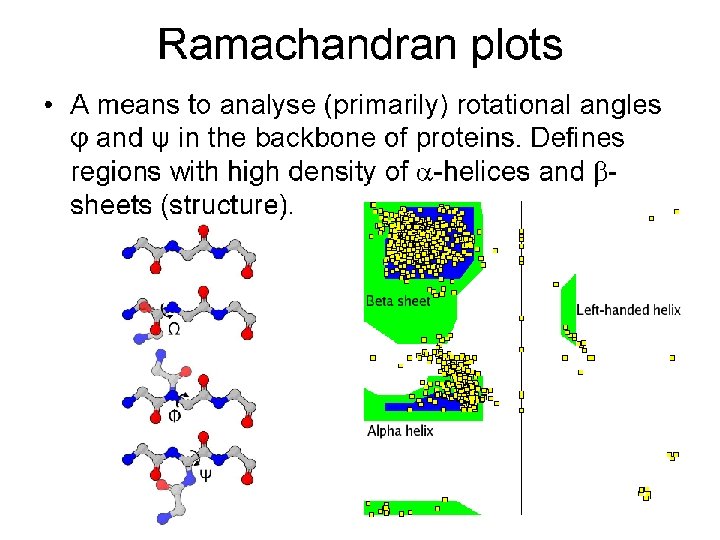
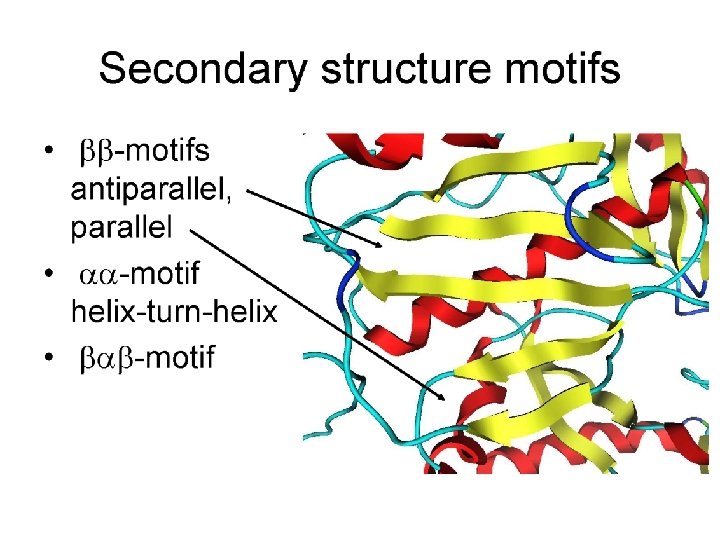
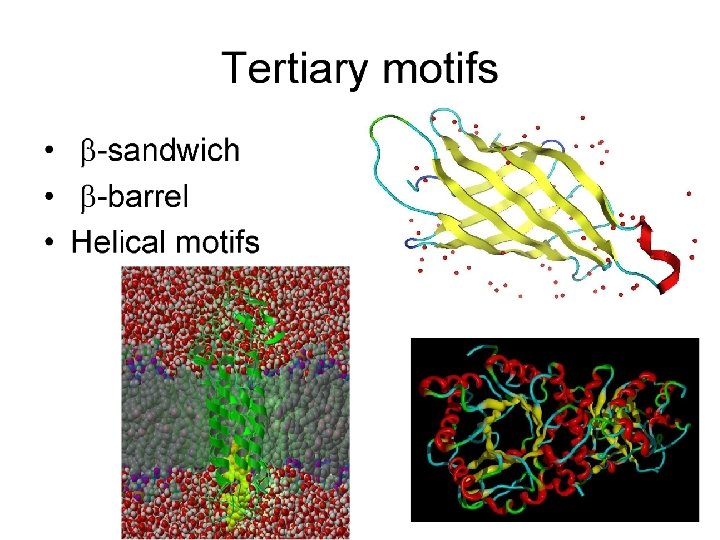
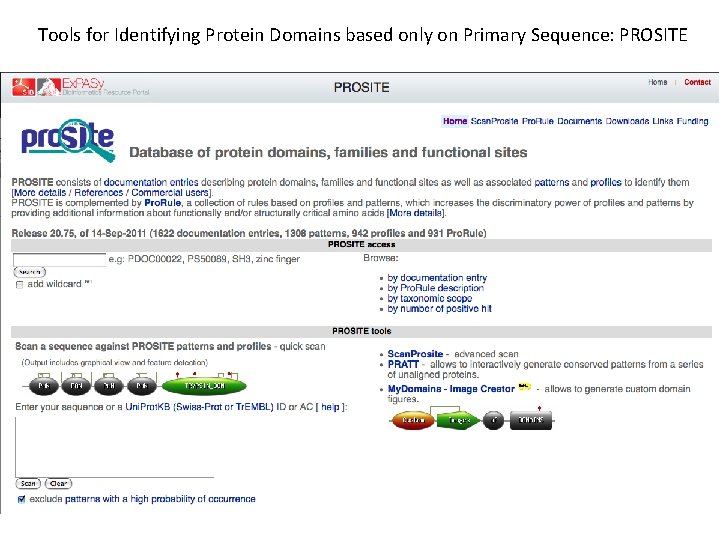
Tools for Identifying Protein Domains based only on Primary Sequence: PROSITE

What is the local sequence preference: ahelix or b-strand or random?
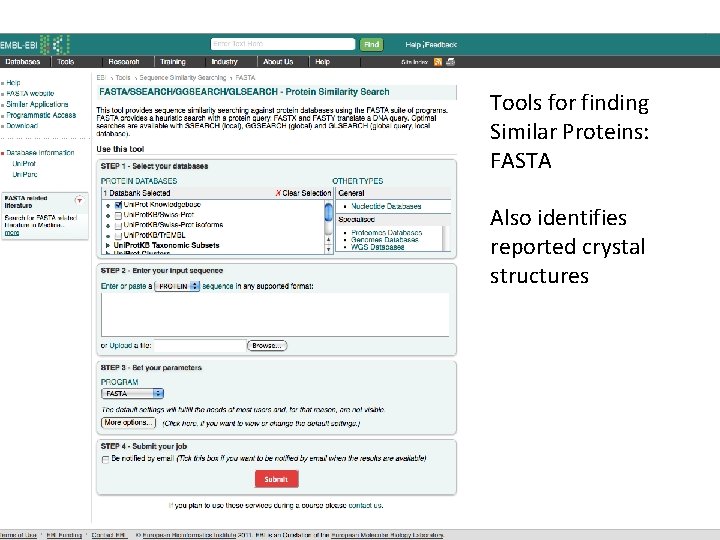
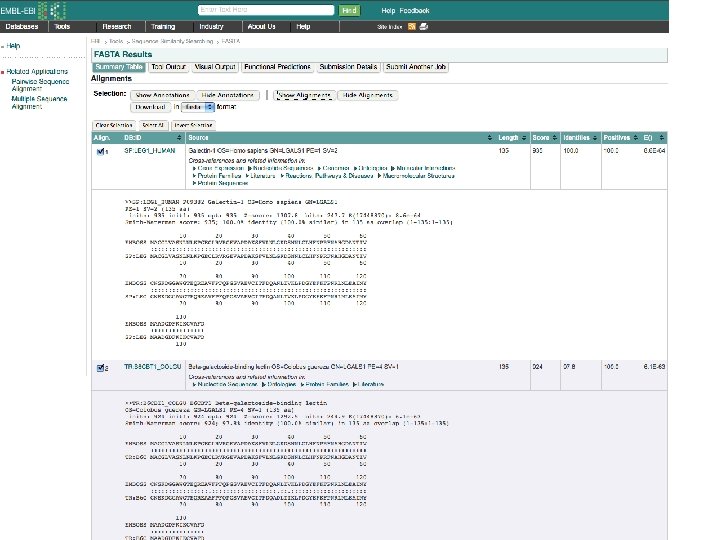
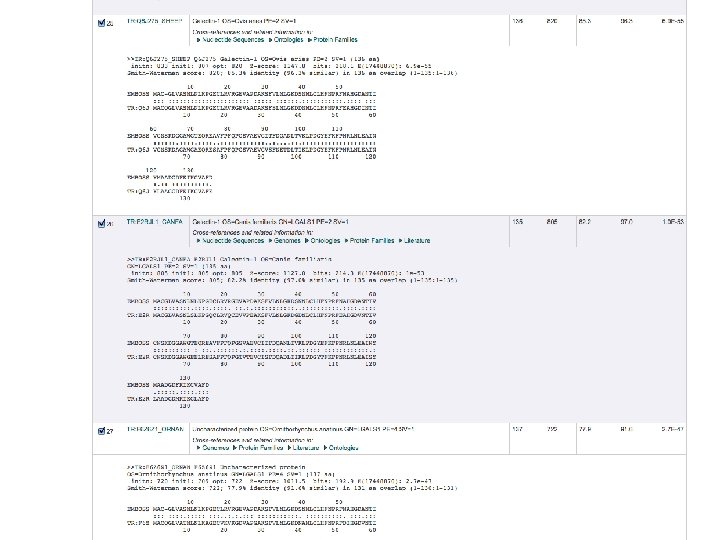
Tools for finding Similar Proteins: FASTA Also identifies reported crystal structures
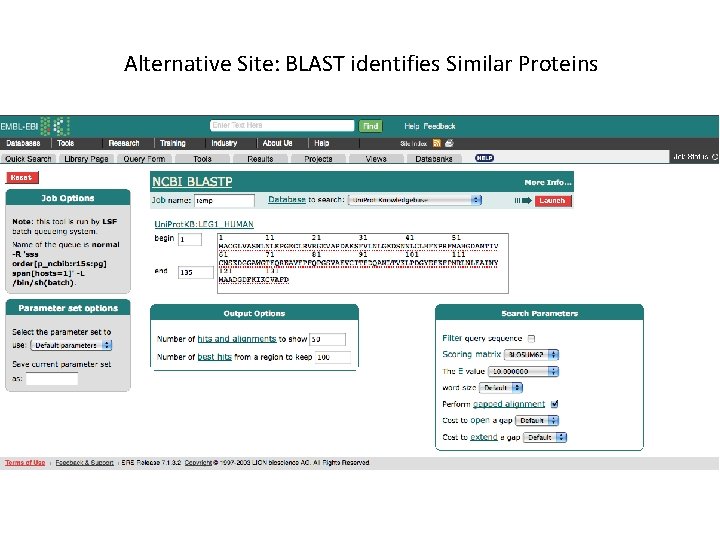
Alternative Site: BLAST identifies Similar Proteins
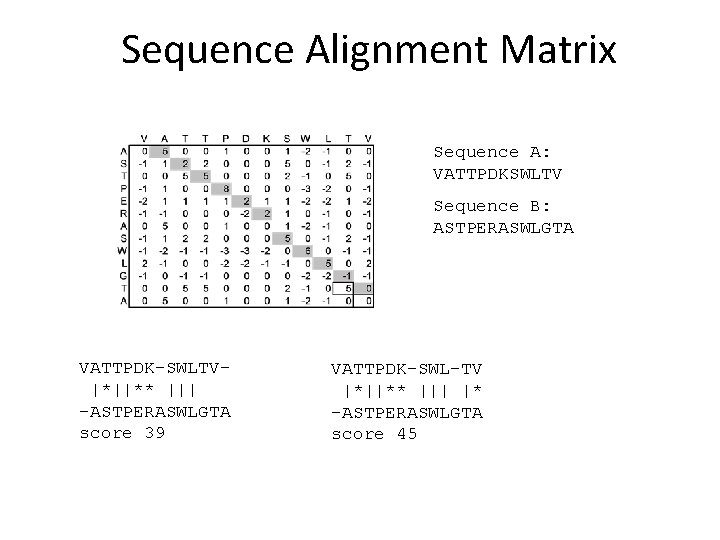
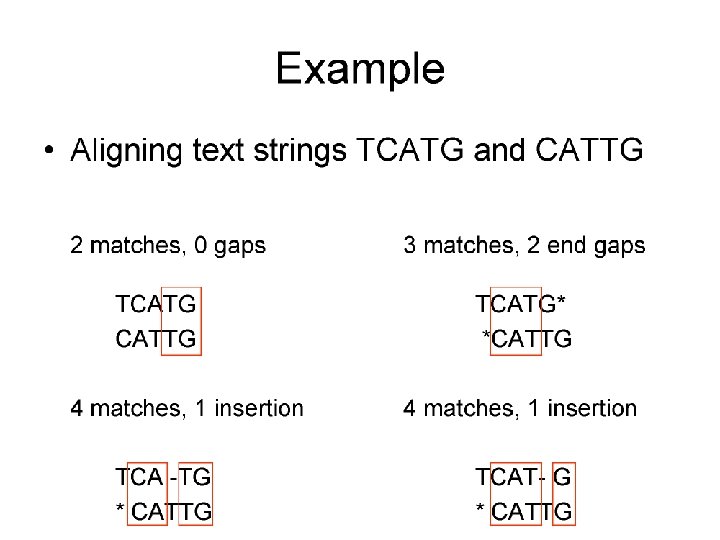
Sequence Alignment Matrix Sequence A: VATTPDKSWLTV Sequence B: ASTPERASWLGTA VATTPDK-SWLTV|*||** ||| -ASTPERASWLGTA score 39 VATTPDK-SWL-TV |*||** ||| |* -ASTPERASWLGTA score 45
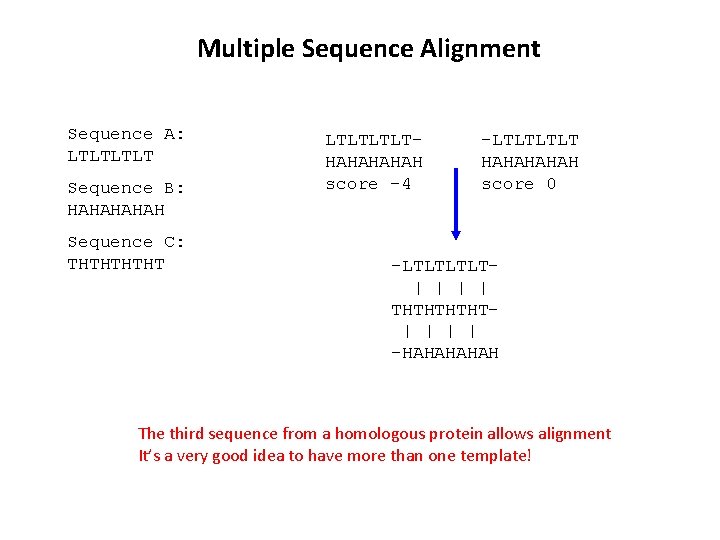
Multiple Sequence Alignment Sequence A: LTLT Sequence B: HAHAH Sequence C: THTHT LTLTHAHAH score -4 -LTLT HAHAH score 0 -LTLT| | THTHT| | -HAHAH The third sequence from a homologous protein allows alignment It’s a very good idea to have more than one template!
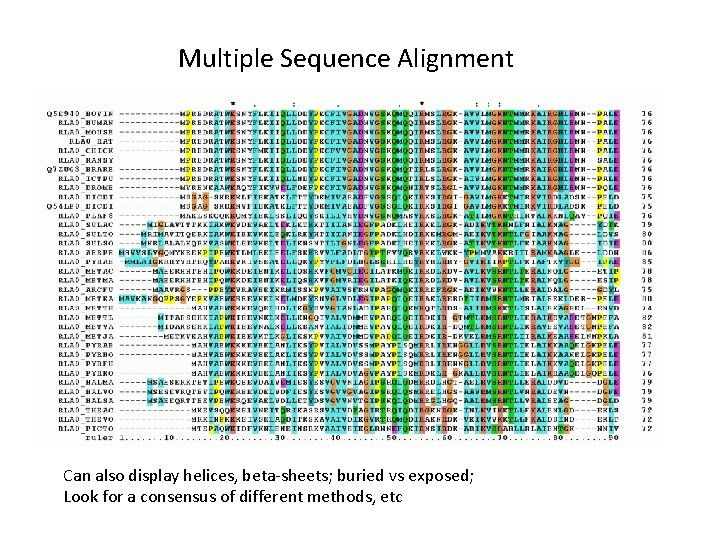
Multiple Sequence Alignment Can also display helices, beta-sheets; buried vs exposed; Look for a consensus of different methods, etc
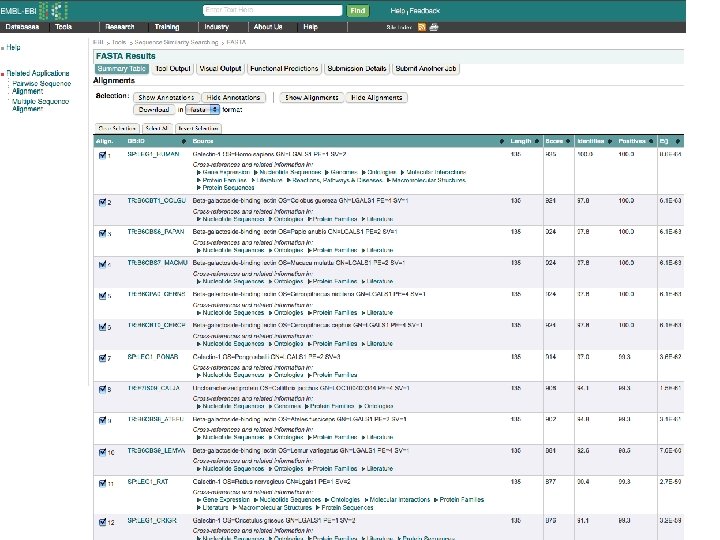
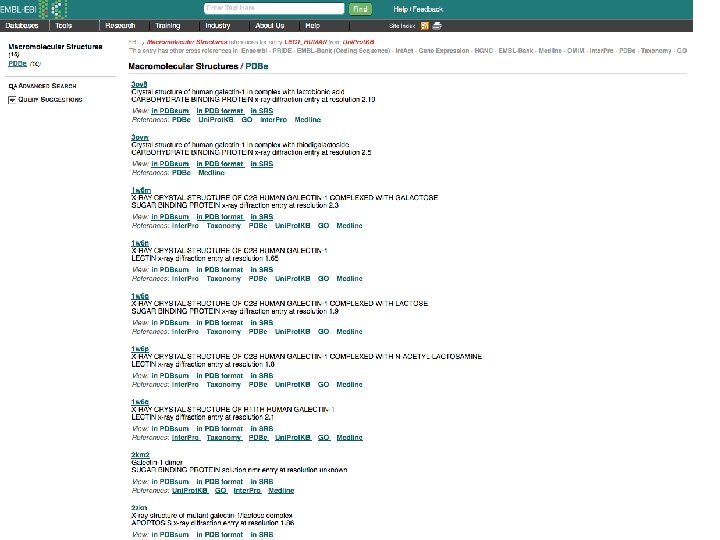

Conclusions Bioinformatics Tools: Can be based on Statistical Analysis, or Scoring, or Physics Sequence matching is the realm of computer science more than chemistry/biochemistry Biochemistry/Chemistry is fundamental to 3 D structure prediction
- Slides: 26