Protein Processing in the Endoplasmic Reticulum Phyllis Hanson



- Slides: 45

Protein Processing in the Endoplasmic Reticulum Phyllis Hanson Cell Biology Dept. , Cancer Res Bldg 4625 phanson 22@wustl. edu 9/8/15

Outline • ER morphology • Protein folding • What happens when protein folding fails – ERAD – UPR • What happens when protein folding is successful – ER exit via COPII vesicles

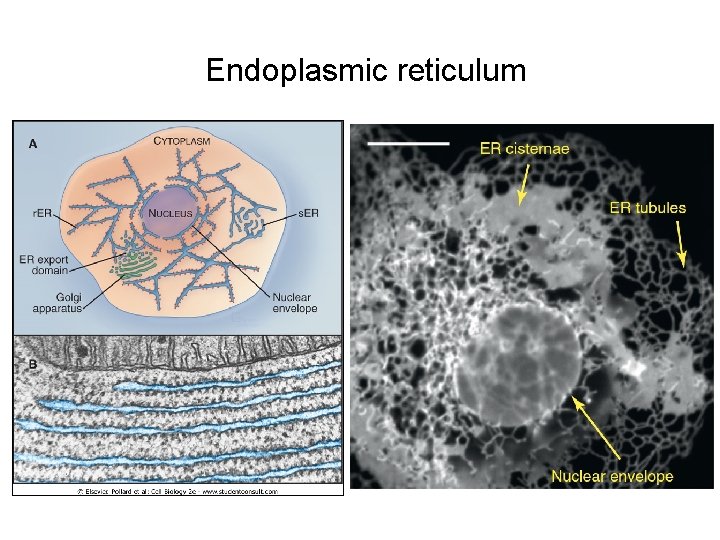
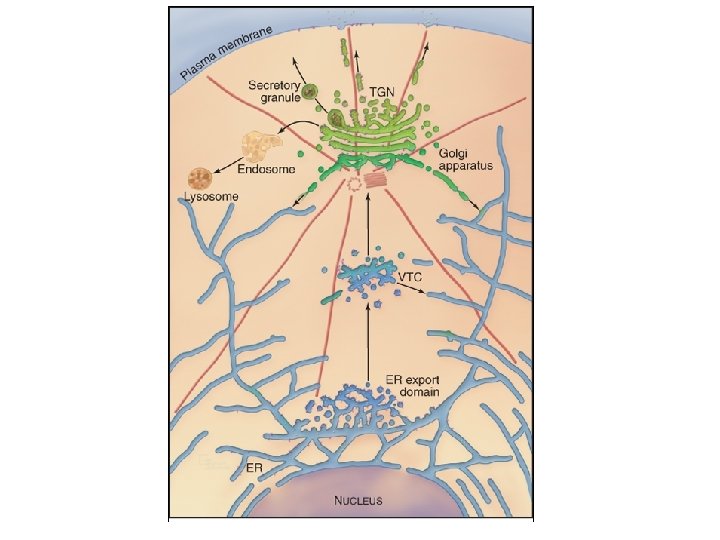
Endoplasmic reticulum

Subdomains of the ER • Rough ER (mostly ER sheets or cisternae) – – • Protein translocation Protein folding and oligomerization Carbohydrate addition ER degradation ] About 1/3 of cellular protein transits through the ER Smooth ER (mostly ER tubules) – Lipid metabolism – Calcium release – Detoxification • ER exit sites (a. k. a. transitional ER) - export of proteins and lipids into the • • Nuclear envelope secretory pathway, marked by COPII coat ER contact zones - transport of lipids, contact with other organelles – Nuclear pores – Chromatin anchoring

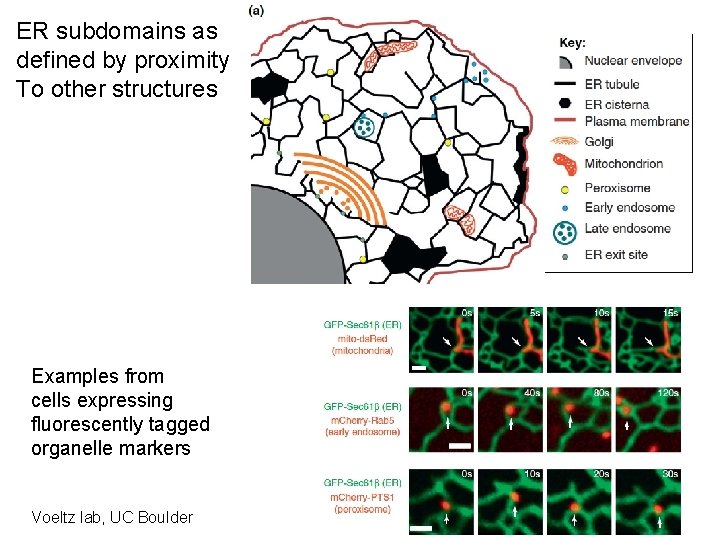
ER subdomains as defined by proximity To other structures Examples from cells expressing fluorescently tagged organelle markers Voeltz lab, UC Boulder

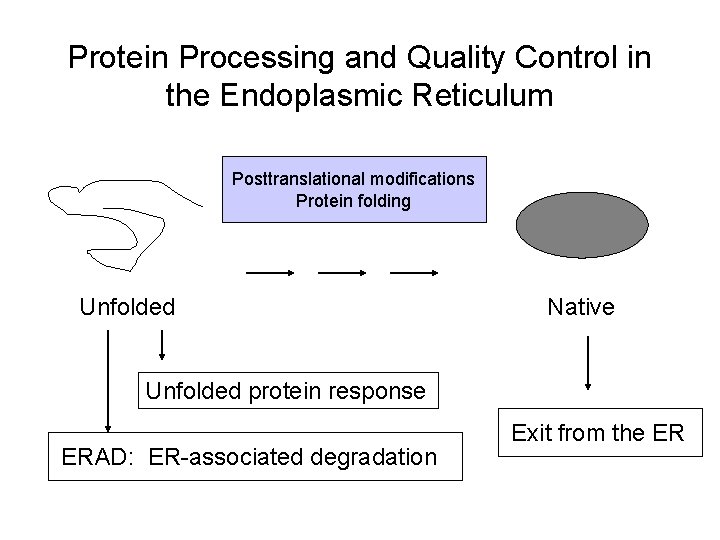
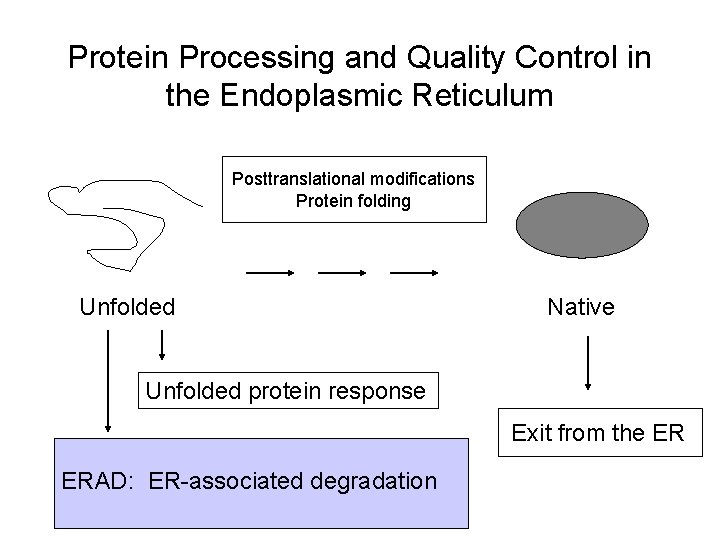
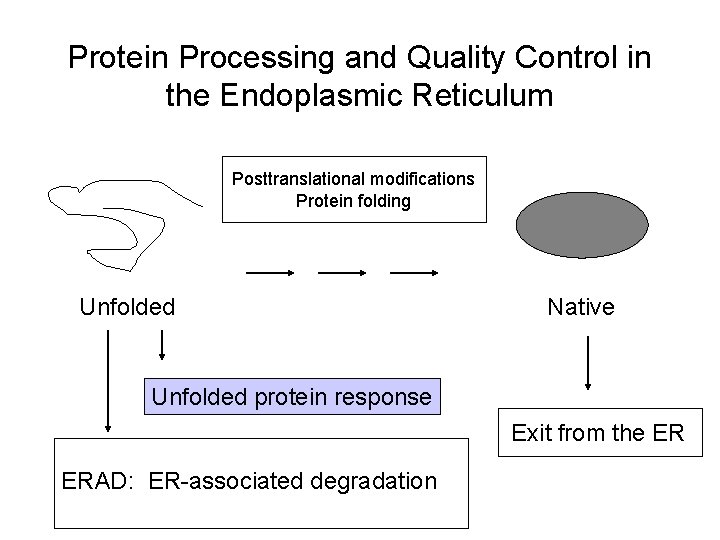
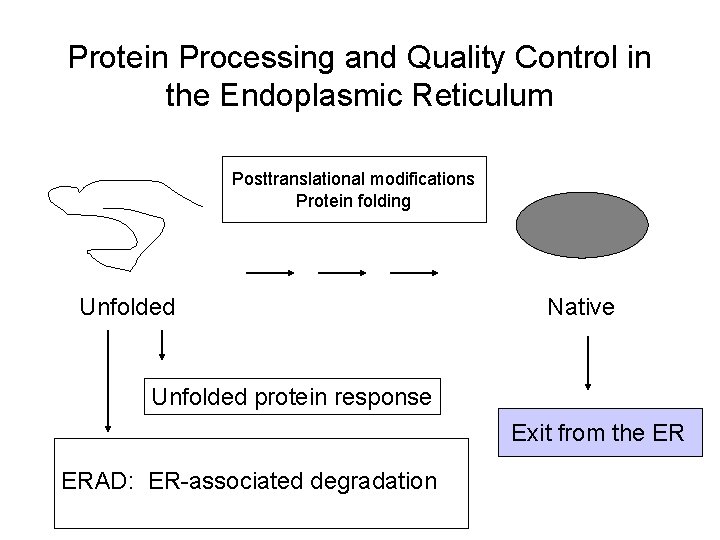
Protein Processing and Quality Control in the Endoplasmic Reticulum Posttranslational modifications Protein folding Unfolded Native Unfolded protein response ERAD: ER-associated degradation Exit from the ER

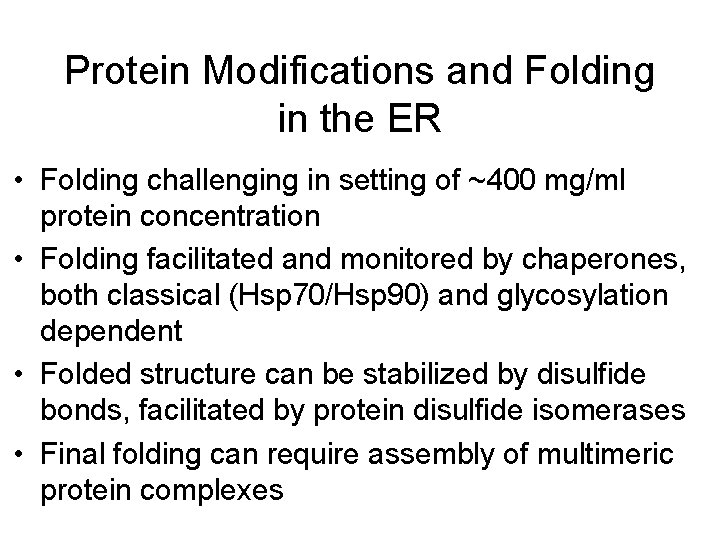
Protein Modifications and Folding in the ER • Folding challenging in setting of ~400 mg/ml protein concentration • Folding facilitated and monitored by chaperones, both classical (Hsp 70/Hsp 90) and glycosylation dependent • Folded structure can be stabilized by disulfide bonds, facilitated by protein disulfide isomerases • Final folding can require assembly of multimeric protein complexes

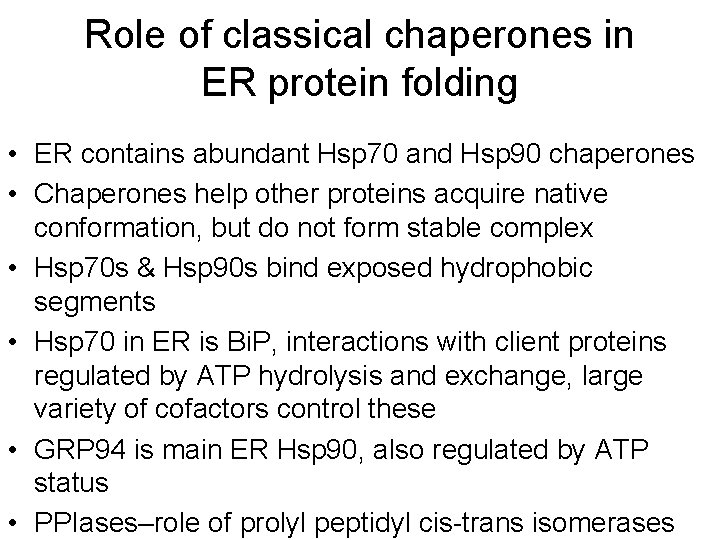
Role of classical chaperones in ER protein folding • ER contains abundant Hsp 70 and Hsp 90 chaperones • Chaperones help other proteins acquire native conformation, but do not form stable complex • Hsp 70 s & Hsp 90 s bind exposed hydrophobic segments • Hsp 70 in ER is Bi. P, interactions with client proteins regulated by ATP hydrolysis and exchange, large variety of cofactors control these • GRP 94 is main ER Hsp 90, also regulated by ATP status • PPIases–role of prolyl peptidyl cis-trans isomerases

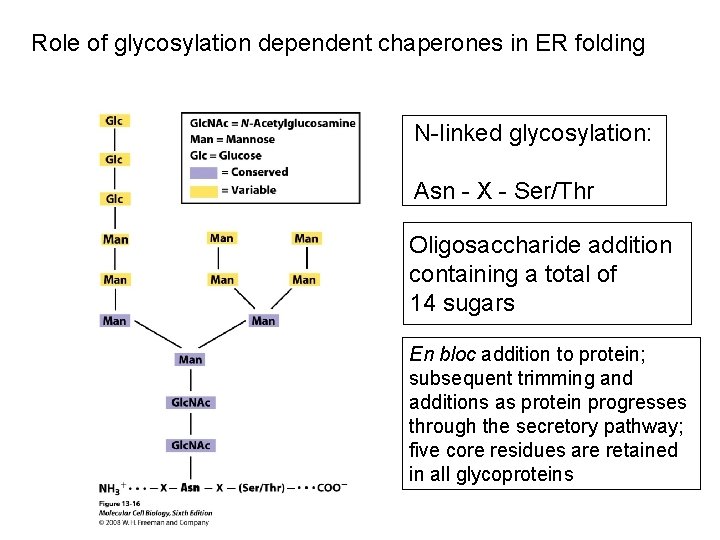
Role of glycosylation dependent chaperones in ER folding N-linked glycosylation: Asn - X - Ser/Thr Oligosaccharide addition containing a total of 14 sugars En bloc addition to protein; subsequent trimming and additions as protein progresses through the secretory pathway; five core residues are retained in all glycoproteins

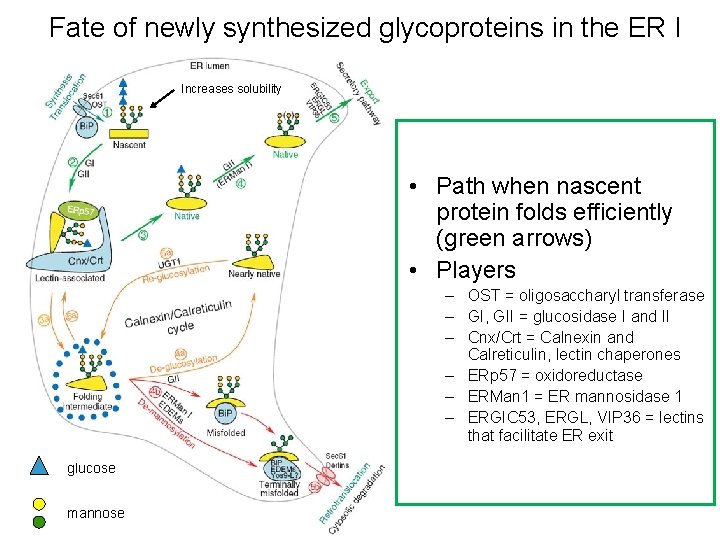
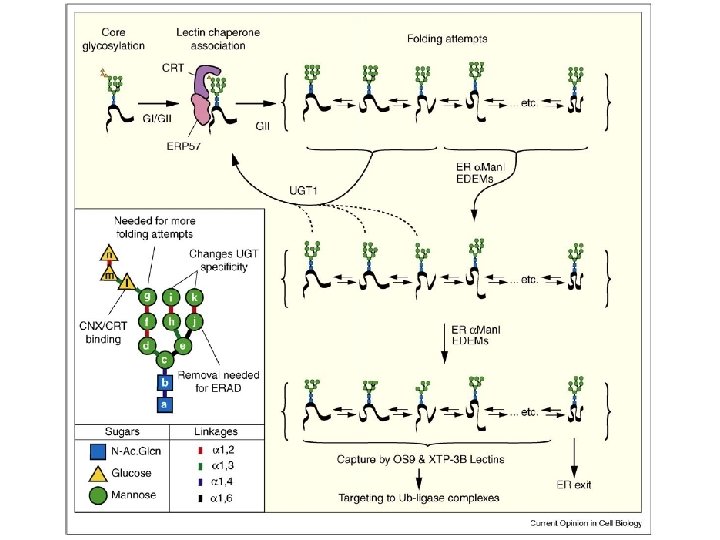
Fate of newly synthesized glycoproteins in the ER I Increases solubility • Path when nascent protein folds efficiently (green arrows) • Players – OST = oligosaccharyl transferase – GI, GII = glucosidase I and II – Cnx/Crt = Calnexin and Calreticulin, lectin chaperones – ERp 57 = oxidoreductase – ERMan 1 = ER mannosidase 1 – ERGIC 53, ERGL, VIP 36 = lectins that facilitate ER exit glucose mannose

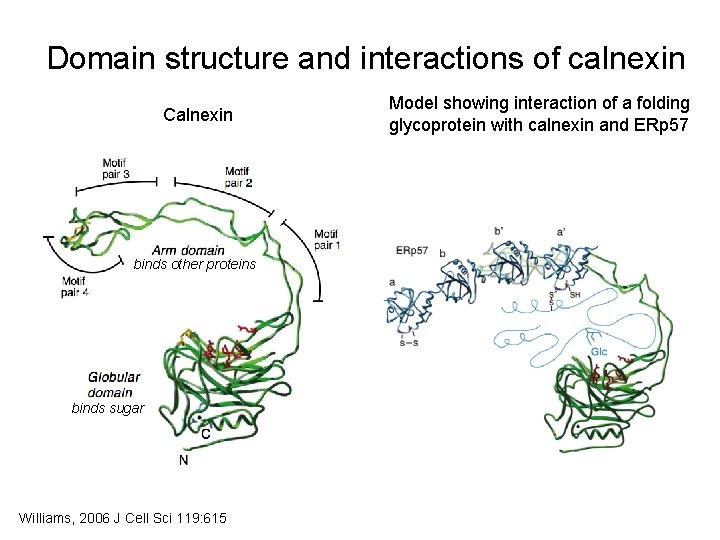
Domain structure and interactions of calnexin Calnexin binds other proteins binds sugar Williams, 2006 J Cell Sci 119: 615 Model showing interaction of a folding glycoprotein with calnexin and ERp 57

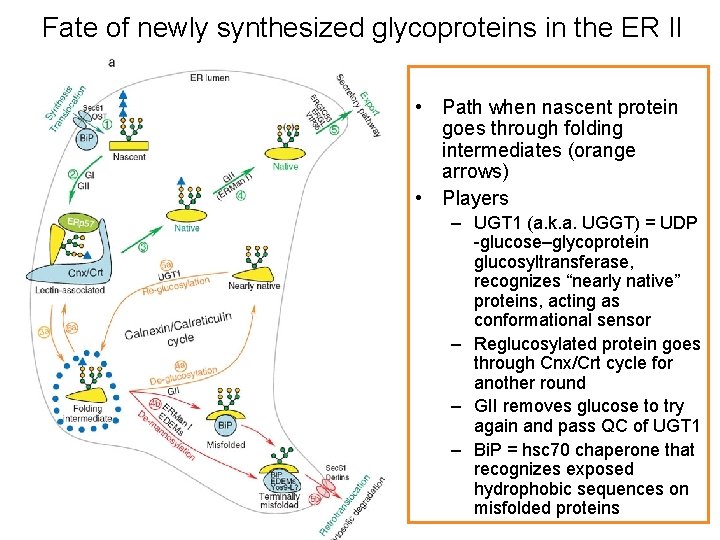
Fate of newly synthesized glycoproteins in the ER II • Path when nascent protein goes through folding intermediates (orange arrows) • Players – UGT 1 (a. k. a. UGGT) = UDP -glucose–glycoprotein glucosyltransferase, recognizes “nearly native” proteins, acting as conformational sensor – Reglucosylated protein goes through Cnx/Crt cycle for another round – GII removes glucose to try again and pass QC of UGT 1 – Bi. P = hsc 70 chaperone that recognizes exposed hydrophobic sequences on misfolded proteins

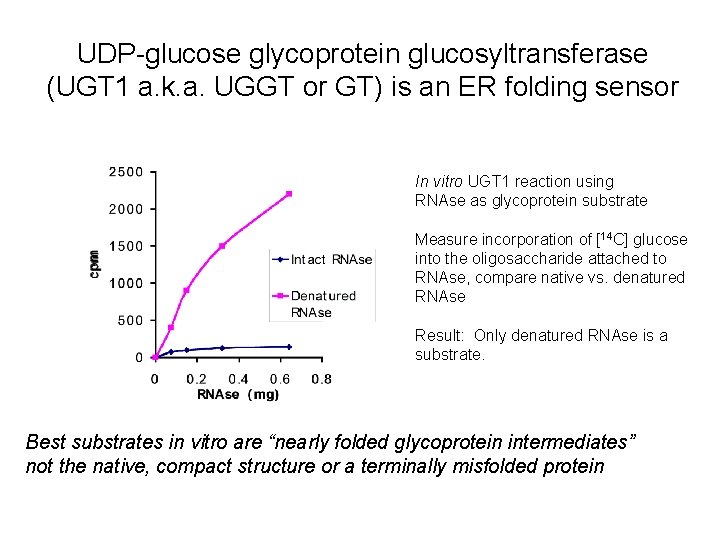
UDP-glucose glycoprotein glucosyltransferase (UGT 1 a. k. a. UGGT or GT) is an ER folding sensor In vitro UGT 1 reaction using RNAse as glycoprotein substrate Measure incorporation of [14 C] glucose into the oligosaccharide attached to RNAse, compare native vs. denatured RNAse Result: Only denatured RNAse is a substrate. Best substrates in vitro are “nearly folded glycoprotein intermediates” not the native, compact structure or a terminally misfolded protein

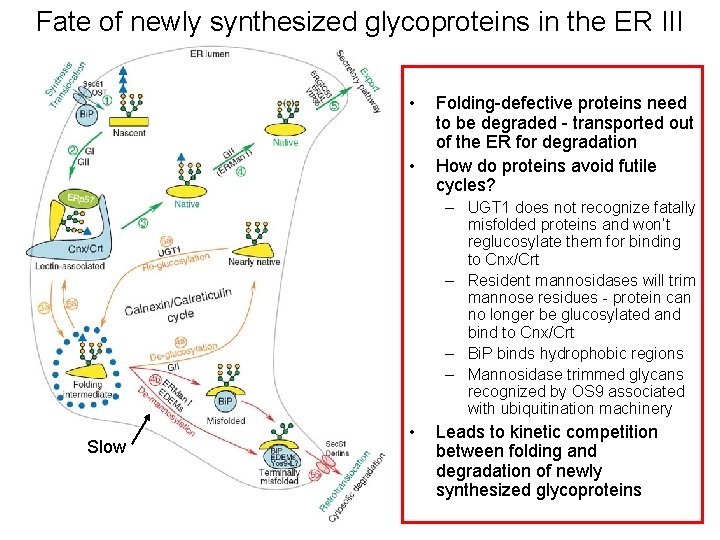
Fate of newly synthesized glycoproteins in the ER III • • Folding-defective proteins need to be degraded - transported out of the ER for degradation How do proteins avoid futile cycles? – UGT 1 does not recognize fatally misfolded proteins and won’t reglucosylate them for binding to Cnx/Crt – Resident mannosidases will trim mannose residues - protein can no longer be glucosylated and bind to Cnx/Crt – Bi. P binds hydrophobic regions – Mannosidase trimmed glycans recognized by OS 9 associated with ubiquitination machinery Slow • Leads to kinetic competition between folding and degradation of newly synthesized glycoproteins


Protein Processing and Quality Control in the Endoplasmic Reticulum Posttranslational modifications Protein folding Unfolded Native Unfolded protein response Exit from the ER ERAD: ER-associated degradation

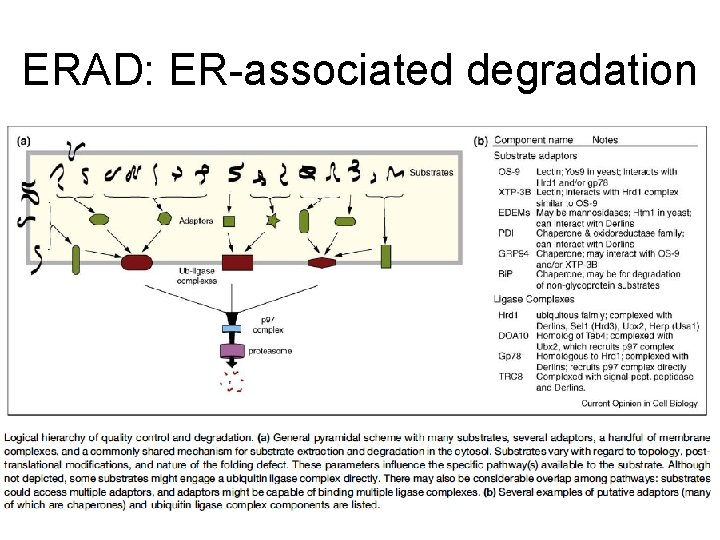
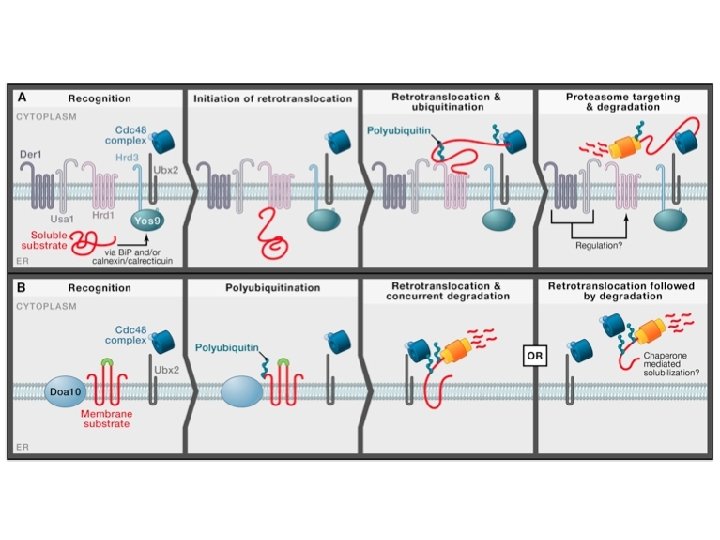
ERAD: ER-associated degradation


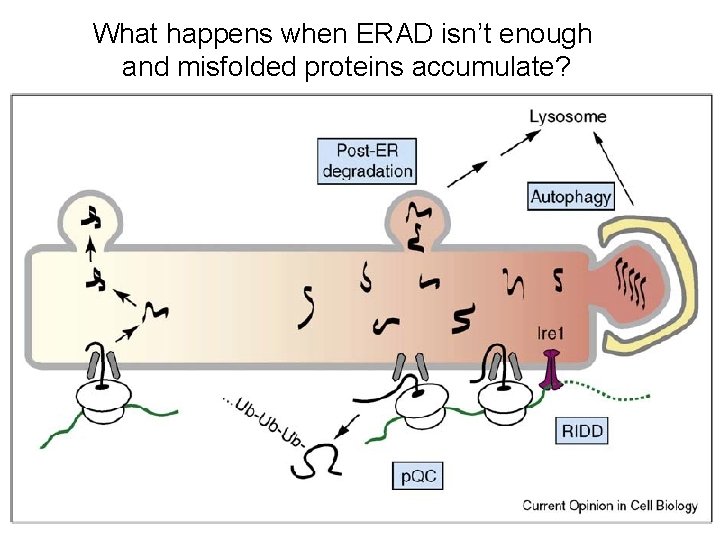
What happens when ERAD isn’t enough and misfolded proteins accumulate?

Protein Processing and Quality Control in the Endoplasmic Reticulum Posttranslational modifications Protein folding Unfolded Native Unfolded protein response Exit from the ER ERAD: ER-associated degradation

Unfolded Protein Response (UPR) • Intracellular signal transduction pathways that mediate communication between ER and nucleus • Activated by accumulation of unfolded proteins in the lumen of the ER • First characterized in yeast • Conserved and more complex in animals, with at least three pathways

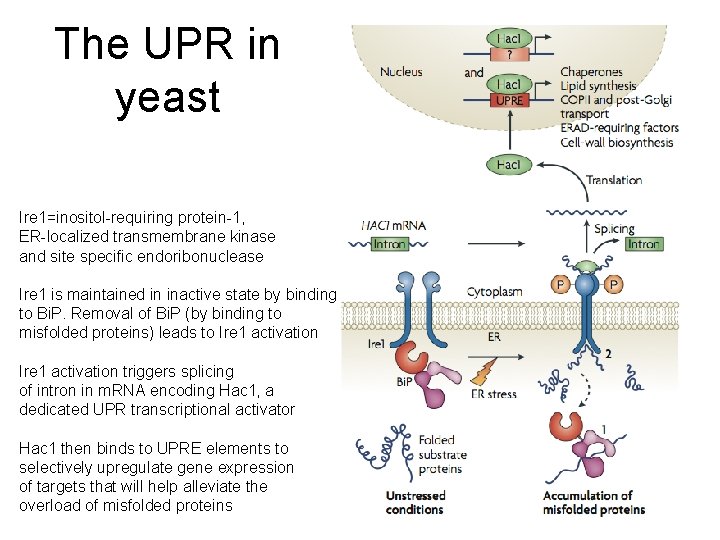
The UPR in yeast Ire 1=inositol-requiring protein-1, ER-localized transmembrane kinase and site specific endoribonuclease Ire 1 is maintained in inactive state by binding to Bi. P. Removal of Bi. P (by binding to misfolded proteins) leads to Ire 1 activation triggers splicing of intron in m. RNA encoding Hac 1, a dedicated UPR transcriptional activator Hac 1 then binds to UPRE elements to selectively upregulate gene expression of targets that will help alleviate the overload of misfolded proteins

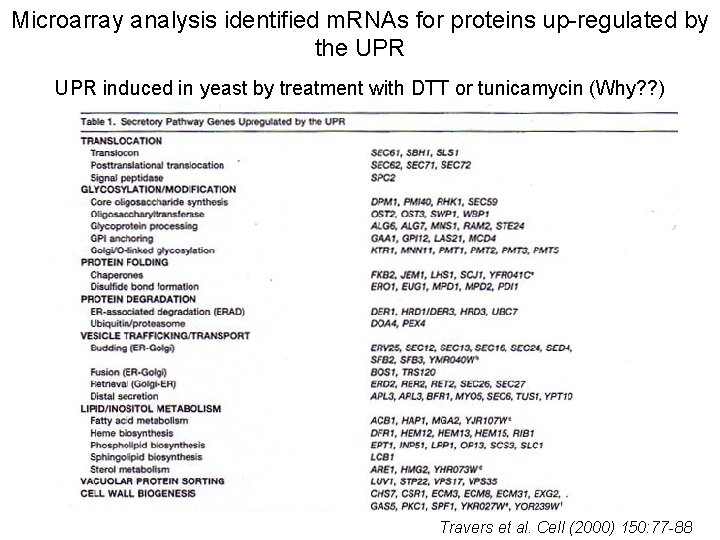
Microarray analysis identified m. RNAs for proteins up-regulated by the UPR induced in yeast by treatment with DTT or tunicamycin (Why? ? ) Travers et al. Cell (2000) 150: 77 -88

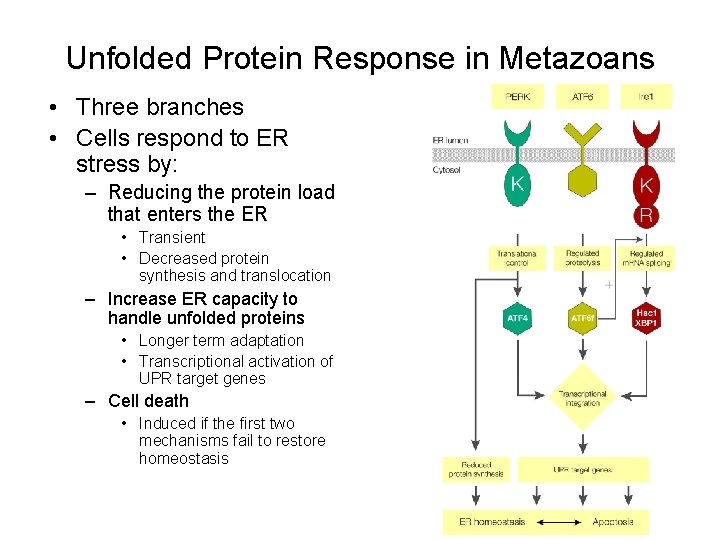
Unfolded Protein Response in Metazoans • Three branches • Cells respond to ER stress by: – Reducing the protein load that enters the ER • Transient • Decreased protein synthesis and translocation – Increase ER capacity to handle unfolded proteins • Longer term adaptation • Transcriptional activation of UPR target genes – Cell death • Induced if the first two mechanisms fail to restore homeostasis

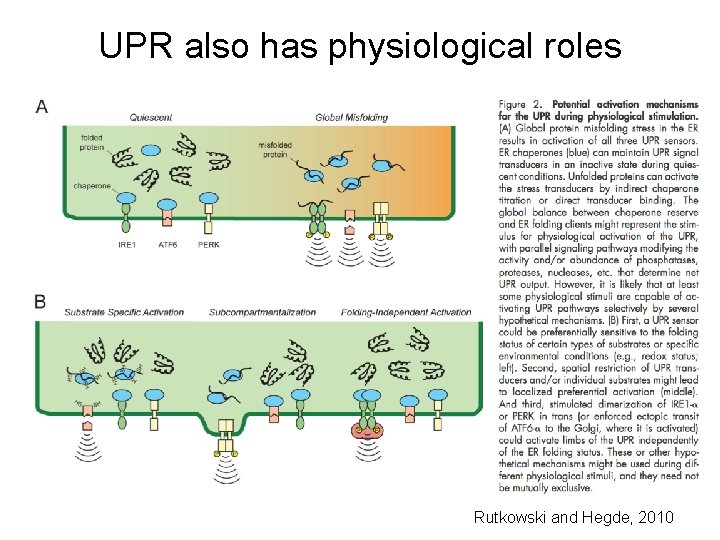
UPR also has physiological roles Rutkowski and Hegde, 2010

Misfolded proteins, ER stress, and disease • Cystic fibrosis transmembrane conductance regulator DF 508 mutation is well studied example (among 100 s known) • Protein could be functional as chloride channel at PM, but does not pass ER QC • Ameliorative strategies include use of chemical chaperones, efforts to modulate specific folding factors, and efforts to adjust overall “proteostasis”

UPR as Achilles heel in Multiple Myeloma? • UPR constitutively activated in setting of uncontrolled immunoglobulin secretion • May make cells particularly vulnerable to drugs that interfere with ER stress response, thereby increasing apoptosis • Proteasome inhibitors now in use, p 97 inhibitors on the way • Others?

Protein Processing and Quality Control in the Endoplasmic Reticulum Posttranslational modifications Protein folding Unfolded Native Unfolded protein response Exit from the ER ERAD: ER-associated degradation


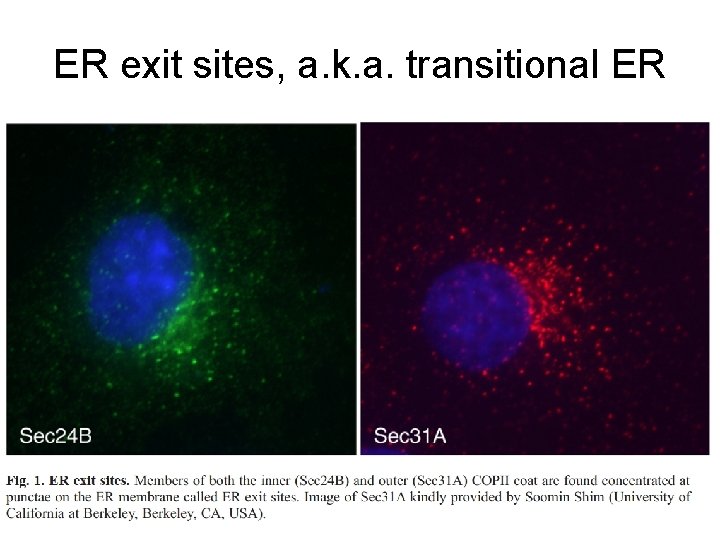
ER exit sites, a. k. a. transitional ER

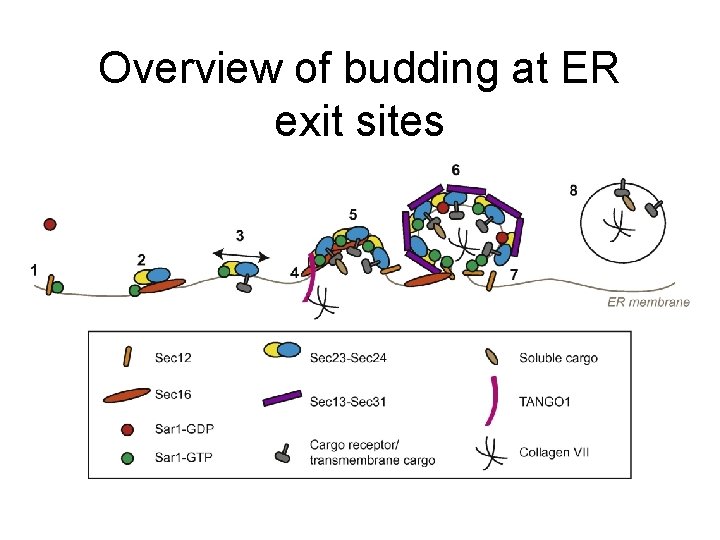
Overview of budding at ER exit sites

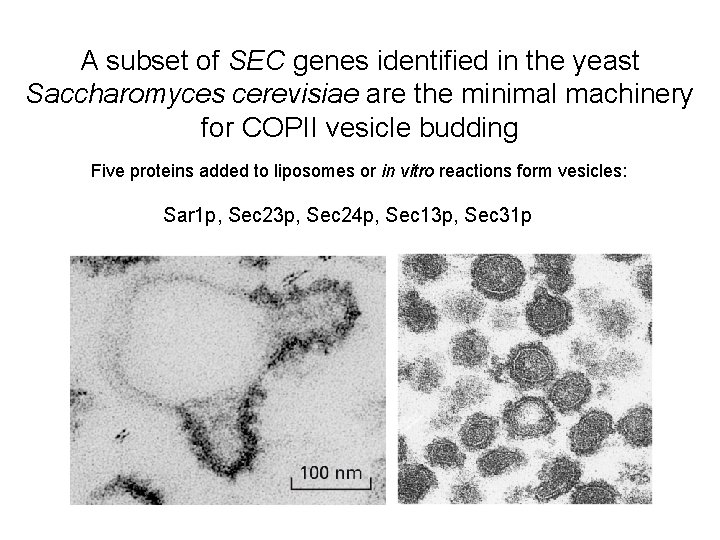
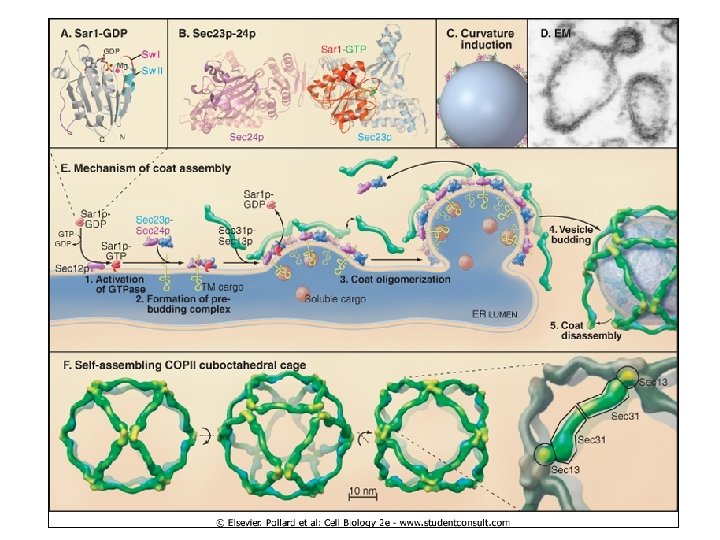
A subset of SEC genes identified in the yeast Saccharomyces cerevisiae are the minimal machinery for COPII vesicle budding Five proteins added to liposomes or in vitro reactions form vesicles: Sar 1 p, Sec 23 p, Sec 24 p, Sec 13 p, Sec 31 p


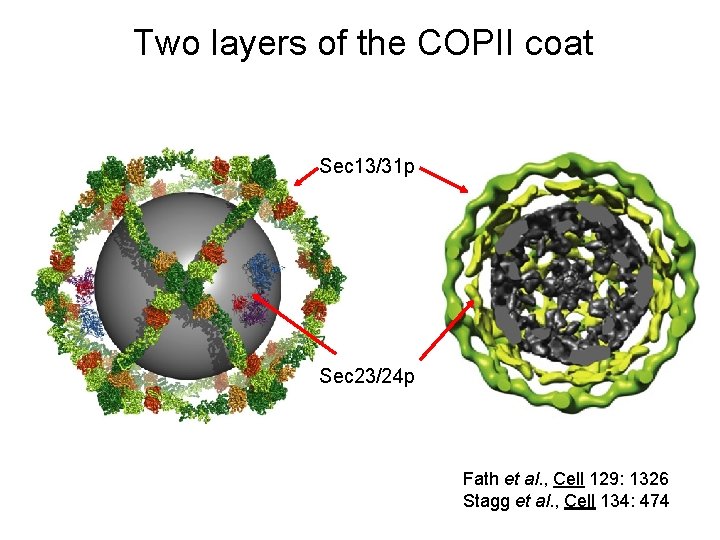
Two layers of the COPII coat Sec 13/31 p Sec 23/24 p Fath et al. , Cell 129: 1326 Stagg et al. , Cell 134: 474

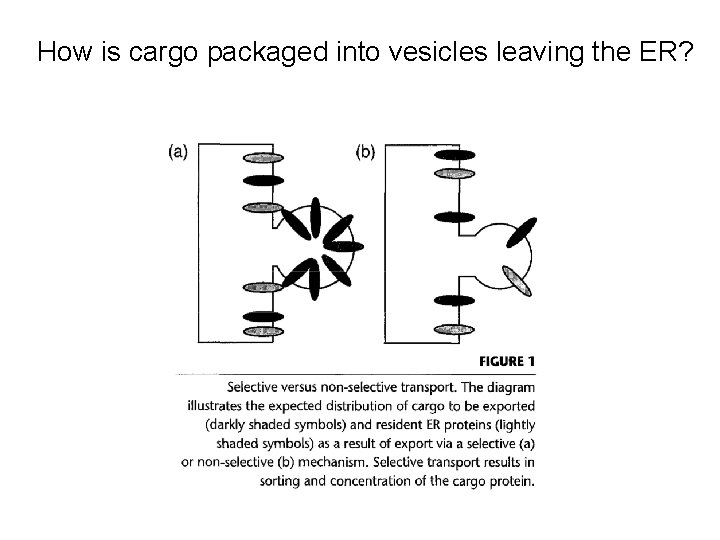
How is cargo packaged into vesicles leaving the ER?


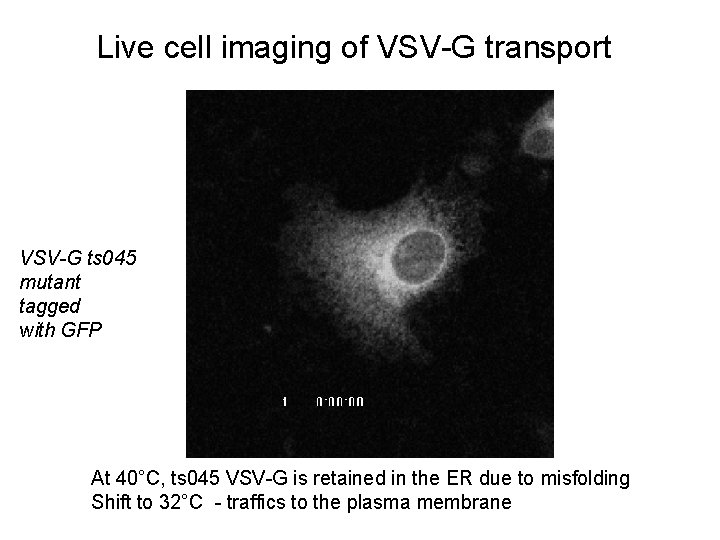
Live cell imaging of VSV-G transport VSV-G ts 045 mutant tagged with GFP At 40°C, ts 045 VSV-G is retained in the ER due to misfolding Shift to 32°C - traffics to the plasma membrane

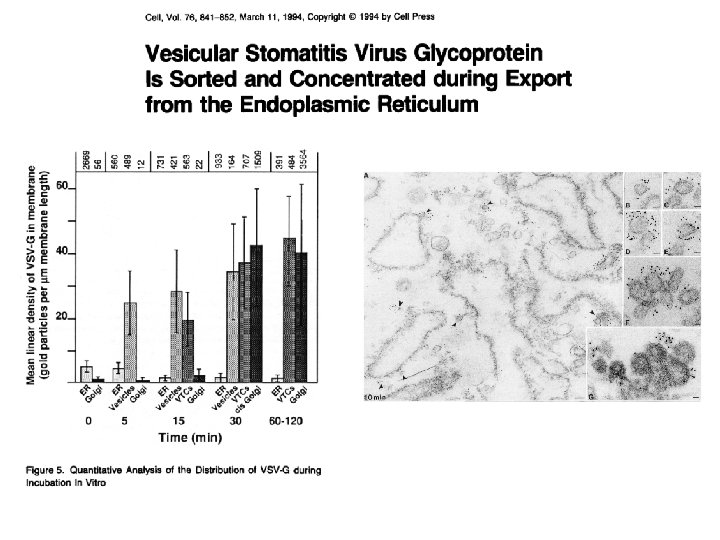
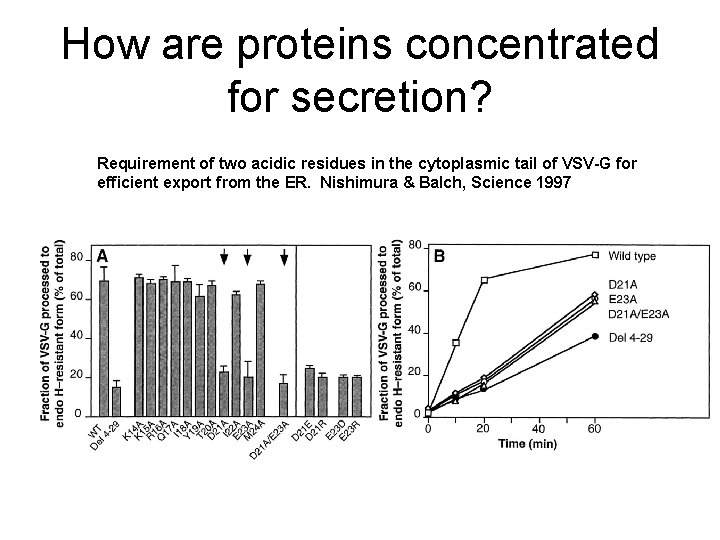
How are proteins concentrated for secretion? Requirement of two acidic residues in the cytoplasmic tail of VSV-G for efficient export from the ER. Nishimura & Balch, Science 1997

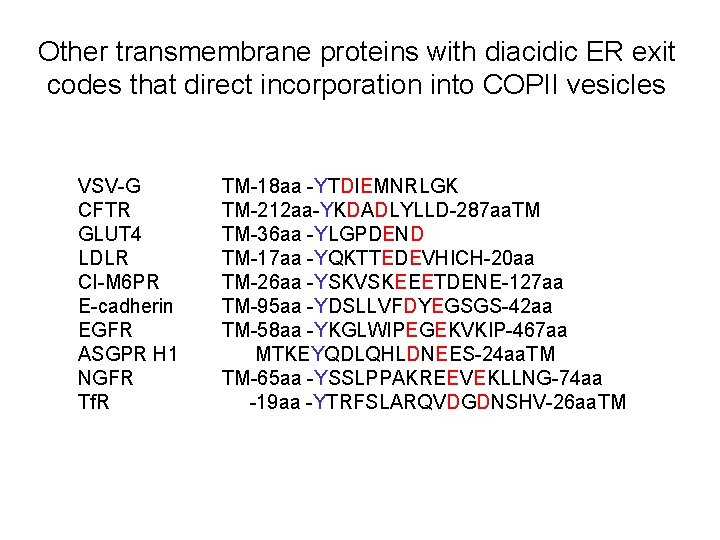
Other transmembrane proteins with diacidic ER exit codes that direct incorporation into COPII vesicles VSV-G CFTR GLUT 4 LDLR CI-M 6 PR E-cadherin EGFR ASGPR H 1 NGFR Tf. R TM-18 aa -YTDIEMNRLGK TM-212 aa-YKDADLYLLD-287 aa. TM TM-36 aa -YLGPDEND TM-17 aa -YQKTTEDEVHICH-20 aa TM-26 aa -YSKVSKEEETDENE-127 aa TM-95 aa -YDSLLVFDYEGSGS-42 aa TM-58 aa -YKGLWIPEGEKVKIP-467 aa MTKEYQDLQHLDNEES-24 aa. TM TM-65 aa -YSSLPPAKREEVEKLLNG-74 aa -19 aa -YTRFSLARQVDGDNSHV-26 aa. TM

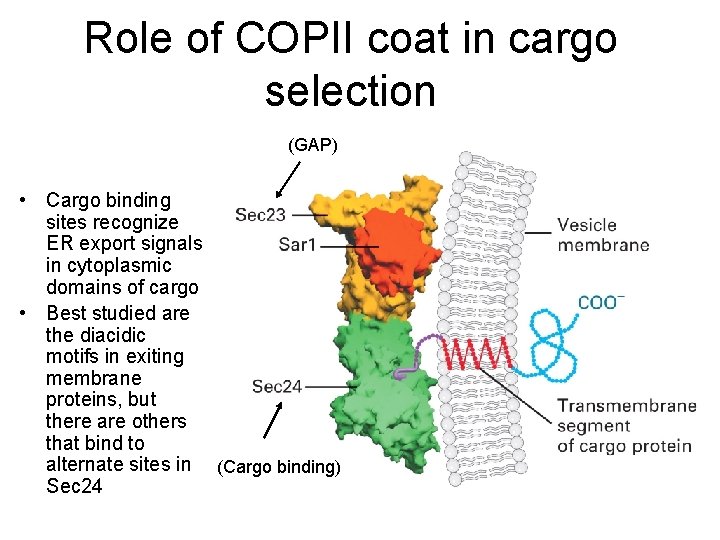
Role of COPII coat in cargo selection (GAP) • Cargo binding sites recognize ER export signals in cytoplasmic domains of cargo • Best studied are the diacidic motifs in exiting membrane proteins, but there are others that bind to alternate sites in (Cargo binding) Sec 24

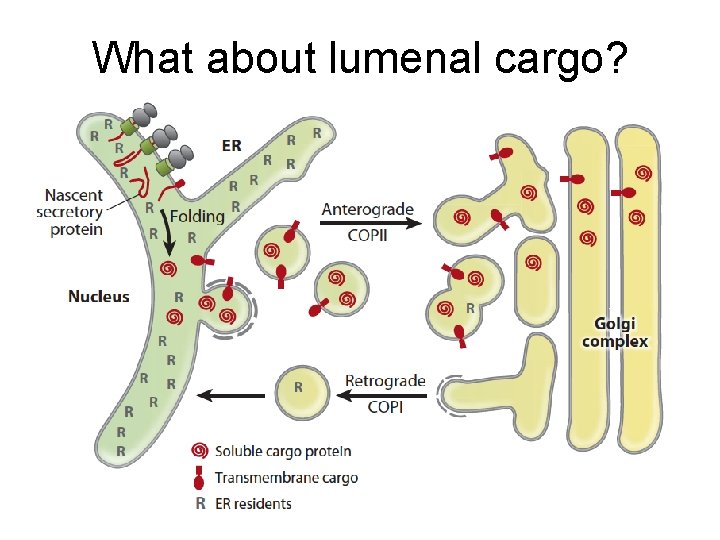
What about lumenal cargo?

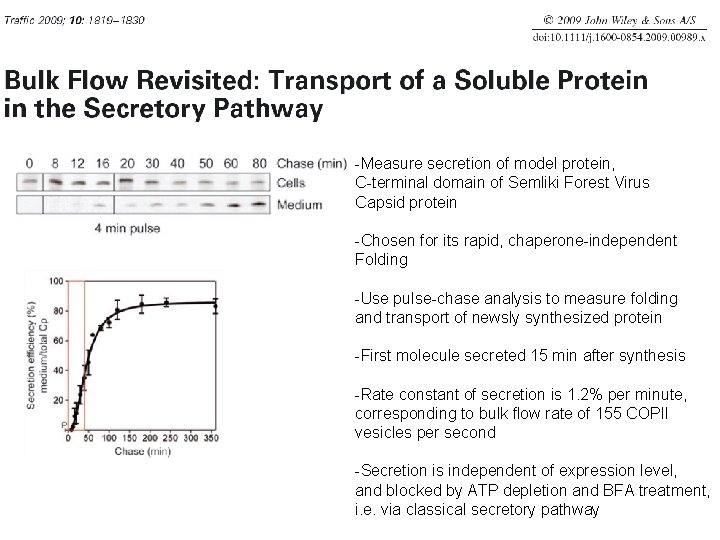
-Measure secretion of model protein, C-terminal domain of Semliki Forest Virus Capsid protein -Chosen for its rapid, chaperone-independent Folding -Use pulse-chase analysis to measure folding and transport of newsly synthesized protein -First molecule secreted 15 min after synthesis -Rate constant of secretion is 1. 2% per minute, corresponding to bulk flow rate of 155 COPII vesicles per second -Secretion is independent of expression level, and blocked by ATP depletion and BFA treatment, i. e. via classical secretory pathway

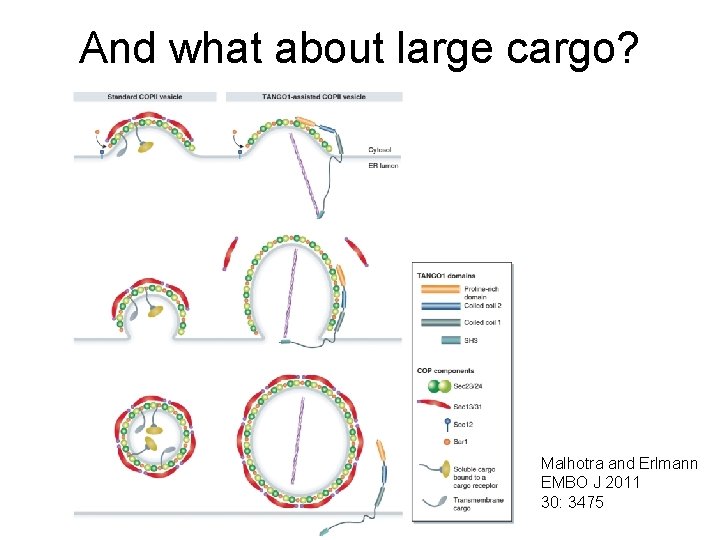
And what about large cargo? Malhotra and Erlmann EMBO J 2011 30: 3475

Next lectures: Thursday: What happens when ER proteins do escape Mechanism of membrane fusion Secretory pathway organelles and trafficking Tuesday: Endocytic pathways and organelles

Reading • Lodish, 7 th edition. Relevant sections of chapters 13 and 14 OR • Pollard, 2 nd edition, Chapter 20, pp. 355 -360 • REVIEW TO READ: “Cleaning up: ERassociated degradation to the rescue”. JL Brodsky, Cell 151: 1163 -1167 • Others of interest on website