Protein Methods Andy Howard Introductory Biochemistry Fall 2013
Protein Methods Andy Howard Introductory Biochemistry, Fall 2013 10 September 2013 Protein Methods and Functions 09/10/2013

Protein methods n Today we’ll finish our discussion about how we learn about proteins 09/10/2013 Protein Methods and Functions p. 2 of 69

Plans for Today n n Purification Methods Structure methods n n n Crystallography NMR Cryo. EM CD, Spectroscopy Scattering, MS 09/10/2013 Protein Methods and Functions p. 3 of 69
Protein Purification n Why do we purify proteins? n n To get a basic idea of function we need to see a protein in isolation from its environment That necessitates purification An instance of reductionist science Full characterization requires a knowledge of the protein’s action in context 09/10/2013 Protein Methods and Functions p. 4 of 69

Salting Out n n Most proteins are less soluble in high salt than in low salt In high salt, water molecules are too busy interacting with the primary solute (salt) to pay much attention to the secondary solute (protein) Various proteins differ in the degree to which their solubility disappears as [salt] goes up We can separate proteins by their differential solubility in high salt. 09/10/2013 Protein Methods and Functions p. 5 of 69

How to do it n n n Dissolve protein mixture in highly soluble salt like Li 2 SO 4, (NH 4)2 SO 4, Na. Cl Increase [salt] until some proteins precipitate and others don’t You may be able to recover both: n n n The supernatant (get rid of salt; move on) The pellet (redissolve, desalt, move on) Typical salt concentrations > 1 M 09/10/2013 Protein Methods and Functions p. 6 of 69
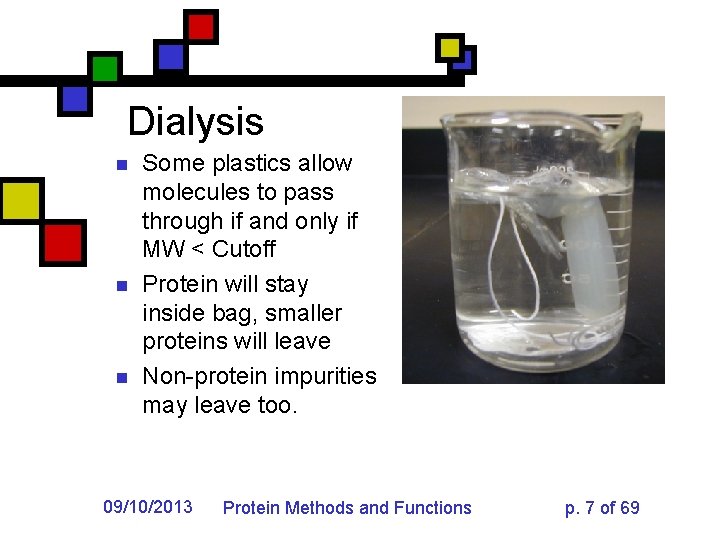
Dialysis n n n Some plastics allow molecules to pass through if and only if MW < Cutoff Protein will stay inside bag, smaller proteins will leave Non-protein impurities may leave too. 09/10/2013 Protein Methods and Functions p. 7 of 69
Gel-filtration chromatography n n n Pass a protein solution through a bead-containing medium at low pressure Beads retard small molecules Beads don’t retard bigger molecules Can be used to separate proteins of significantly different sizes Suitable for preparative work 09/10/2013 Protein Methods and Functions p. 8 of 69
Ion-exchange chromatography n n Charged species affixed to column Phosphonates (-) retard (+)charged proteins: Cation exchange Quaternary ammonium salts (+) retard (-)charged proteins: Anion exchange Separations facilitated by adjusting p. H 09/10/2013 Protein Methods and Functions p. 9 of 69
Affinity chromatography n n n Stationary phase contains a species that has specific favorable interaction with the protein we want DNA-binding protein specific to AGCATGCT: bind AGCATGCT to a column, and the protein we want will stick; every other protein falls through Often used to purify antibodies by binding the antigen to the column 09/10/2013 Protein Methods and Functions p. 10 of 69
Metal-ion affinity chromatography n n Immobilize a metal ion, e. g. Ni, to the column material Proteins with affinity to that metal will stick Wash them off afterward with a ligand with an even higher affinity We can engineer proteins to contain the affinity tag: poly-histidine at N- or C-terminus 09/10/2013 Protein Methods and Functions p. 11 of 69
High-performance liquid chromatography n n Many LC separations can happen faster and more effectively under high pressure Works for small molecules Protein application is routine too, both for analysis and purification FPLC is a trademark, but it’s used generically 09/10/2013 Protein Methods and Functions p. 12 of 69
Electrophoresis n n n Separating analytes by charge by subjecting a mixture to a strong electric field Gel electrophoresis: field applied to a semisolid matrix Can be used for charge (directly) or size (indirectly) 09/10/2013 Protein Methods and Functions p. 13 of 69
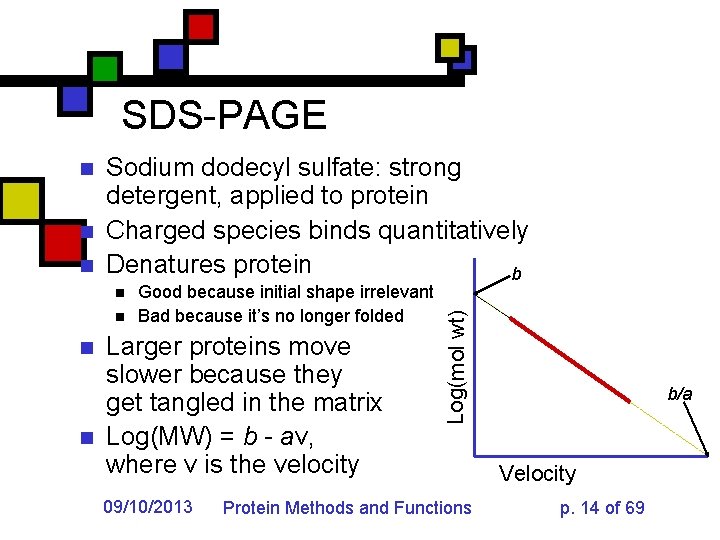
SDS-PAGE n n Sodium dodecyl sulfate: strong detergent, applied to protein Charged species binds quantitatively Denatures protein b n n Good because initial shape irrelevant Bad because it’s no longer folded Larger proteins move slower because they get tangled in the matrix Log(MW) = b - av, where v is the velocity 09/10/2013 Log(mol wt) n Protein Methods and Functions b/a Velocity p. 14 of 69
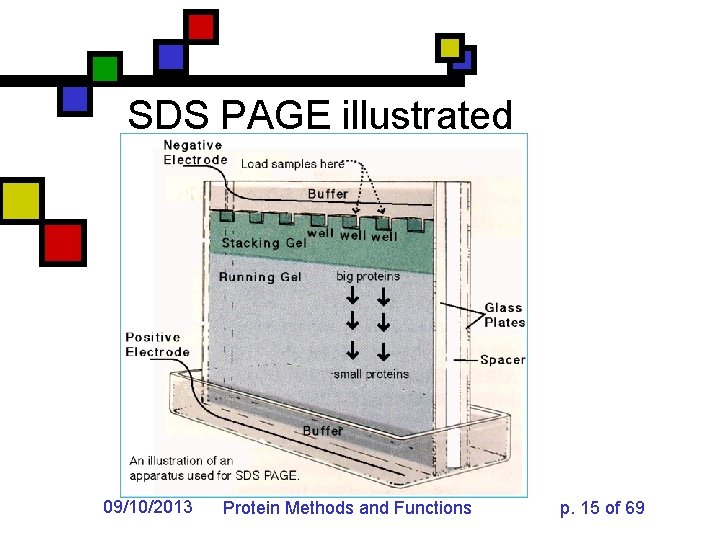
SDS PAGE illustrated 09/10/2013 Protein Methods and Functions p. 15 of 69
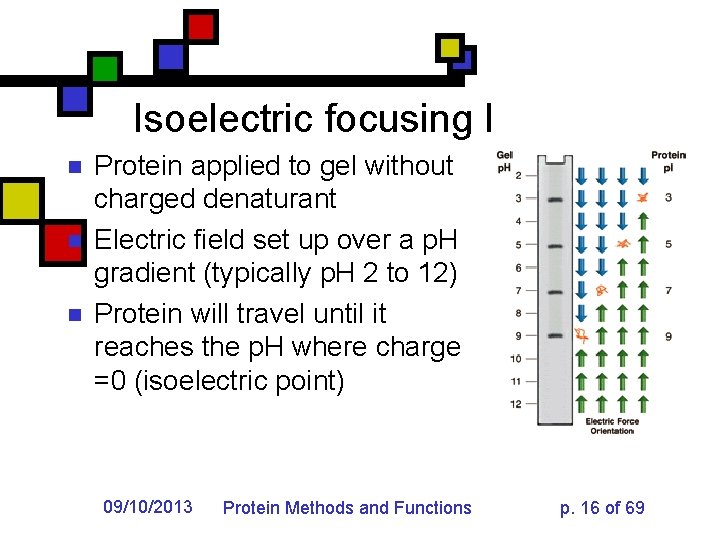
Isoelectric focusing I n n n Protein applied to gel without charged denaturant Electric field set up over a p. H gradient (typically p. H 2 to 12) Protein will travel until it reaches the p. H where charge =0 (isoelectric point) 09/10/2013 Protein Methods and Functions p. 16 of 69

Isoelectric focusing II n n Sensitive to single changes in charge (e. g. asp asn) Can be readily used preparatively with samples that are already semipure 09/10/2013 Protein Methods and Functions p. 17 of 69
Ultraviolet spectroscopy n n n Tyr, trp absorb and fluoresce: abs ~ 280 -274 nm; f = 348 (trp), 303 nm (tyr) Reliable enough to use for estimating protein concentration via Beer’s law (A = Cl ) UV absorption peaks for cofactors in various states are well-understood More relevant for identification of moieties than for structure determination Quenching of fluorescence sometimes provides structural information 09/10/2013 Protein Methods and Functions p. 18 of 69
Mass spectrometry n n Enables calculation of mass to charge ratio for molecules or fragments 1980’s: techniques for aerosolizing proteins were developed so that MS could be applied to proteins n n n Electrospray Matrix-assisted laser desorption ionization Useful analytical tool, alone or along with partial digestion by proteases 09/10/2013 Protein Methods and Functions p. 19 of 69

Warning: Specialty Content! n n n I determine protein structures (and develop methods for determining protein structures) as my own research focus So it’s hard for me to avoid putting a lot of emphasis on this material But today I’m allowed to do that, because it’s one of the stated topics of the day. 09/10/2013 Protein Methods and Functions p. 20 of 69
How do we determine structure? n n We can distinguish between methods that require little prior knowledge (crystallography, NMR, Cryo. EM) and methods that answer specific questions (XAFS, fiber, …) This distinction isn’t entirely clear-cut 09/10/2013 Protein Methods and Functions p. 21 of 69

Crystallography: overview n n Crystals are translationally ordered 3 -D arrays of molecules Conventional solids are usually crystals Proteins have to be coerced into crystallizing … but once they’re crystals, they behave like other crystals, mostly 09/10/2013 Protein Methods and Functions p. 22 of 69
How are protein crystals unusual? n n Aqueous interactions required for crystal integrity: they disintegrate if dried Bigger unit cells (~10 -40 nm, not 0. 2 - 1 nm) Small # of unit cells and static disorder means they don’t scatter terribly well So using them to determine 3 D structures is feasible but difficult 09/10/2013 Protein Methods and Functions p. 23 of 69
Crystal structures: Fourier transforms of diffraction results n Experiment: n n n n Grow crystal, expose it to X-ray Record diffraction spots Rotate through small angle and repeat ~180 times Position of spots tells you size, shape of unit cell Intensity tells you what the contents are We’re using electromagnetic radiation, which behaves like a wave, exp(2 ik • x) Therefore intensity Ihkl = C*|Fhkl|2 09/10/2013 Protein Methods and Functions p. 24 of 69
What are these Fhkl values? n n Fhkl is a complex coefficient in the Fourier transform of the electron density in the unit cell: (r) = (1/V) hkl Fhkl exp(-2 ih • r) Critical point: any single diffraction spot contains information derived from all the atoms in the structure; and any atom contributes to all the diffraction spots 09/10/2013 Protein Methods and Functions p. 25 of 69
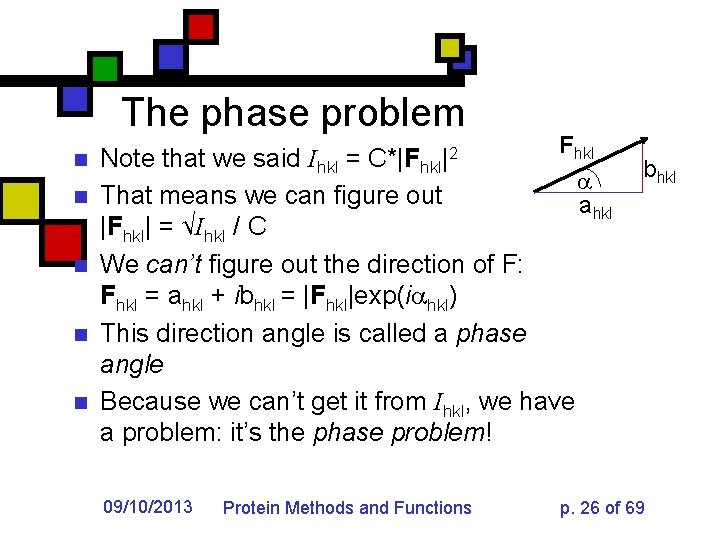
The phase problem n n n F hkl Note that we said Ihkl = C*|Fhkl|2 That means we can figure out ahkl |Fhkl| = √Ihkl / C We can’t figure out the direction of F: Fhkl = ahkl + ibhkl = |Fhkl|exp(i hkl) This direction angle is called a phase angle Because we can’t get it from Ihkl, we have a problem: it’s the phase problem! 09/10/2013 Protein Methods and Functions bhkl p. 26 of 69
Solving the phase problem n n Small molecules: use relationships among intensities of related spots to calculate phases Big molecules: n n Add heavy atom to protein and measure the changes to the diffraction Change the wavelength and measure how that affects the diffraction Start with a model based on a related structure and calculate how to fit that structure into the new unit cell After you do that, it’s usually straightforward to calculate αhkl -> ρ(r) 09/10/2013 Protein Methods and Functions p. 27 of 69

What can we learn? n n n Electron density map + sequence we can determine the positions of all the non-H atoms in the protein—maybe! Best resolution possible: Dmin = / 2 Often the crystal doesn’t diffract that well, so Dmin is larger— 1. 5Å, 2. 5Å, worse Dmin ~ 2. 5Å tells us where backbone and most side-chain atoms are Dmin ~ 1. 2Å: all protein non-H atoms, most solvent, some disordered atoms; some H’s 09/10/2013 Protein Methods and Functions p. 28 of 69
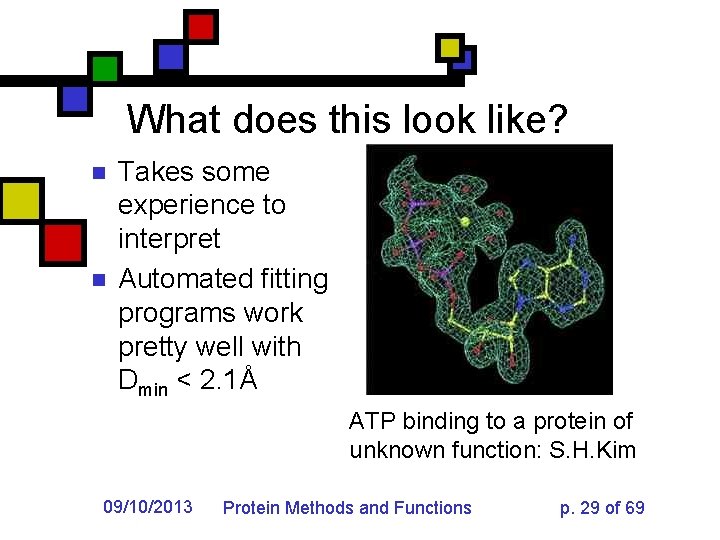
What does this look like? n n Takes some experience to interpret Automated fitting programs work pretty well with Dmin < 2. 1Å ATP binding to a protein of unknown function: S. H. Kim 09/10/2013 Protein Methods and Functions p. 29 of 69
How’s the field changing? n n n 1990: all structures done by professionals Now: many biochemists and molecular biologists are launching their own structure projects as part of broader functional studies Fearless prediction: by 2020: n n crystallographers will be either technicians or methods developers Most structures will be determined by cell biologists & molecular biologists 09/10/2013 Protein Methods and Functions p. 30 of 69
Macromolecular NMR n n n NMR is a mature field Depends on resonant interaction between EM fields and unpaired nucleons (1 H, 15 N, 31 S) Raw data yield interatomic distances Conventional spectra of proteins are too muddy to interpret Multi-dimensional (2 -4 D) techniques: initial resonances coupled with additional ones 09/10/2013 Protein Methods and Functions p. 31 of 69
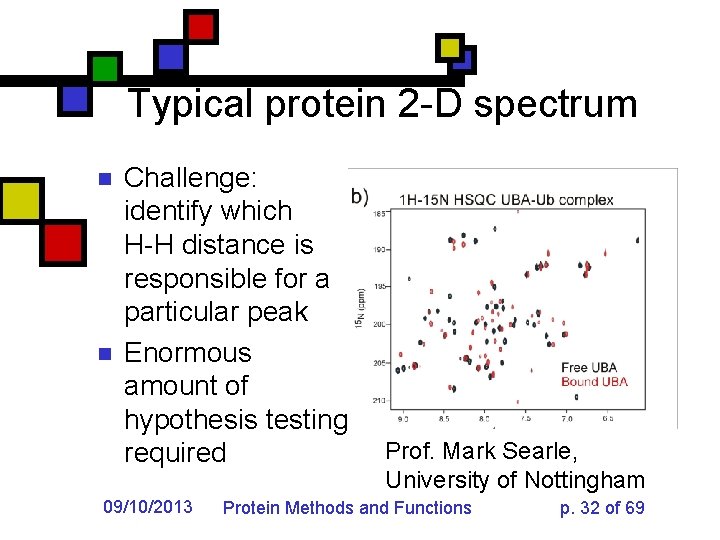
Typical protein 2 -D spectrum n n Challenge: identify which H-H distance is responsible for a particular peak Enormous amount of hypothesis testing required 09/10/2013 Prof. Mark Searle, University of Nottingham Protein Methods and Functions p. 32 of 69
Results n n Often there’s a family of structures that satisfy the NMR data equally well Can be portrayed as a series of threads tied down at unambiguous assignments They portray the protein’s structure in solution The ambiguities partly represent real molecular diversity; but they also represent atoms that area in truth well-defined, but the NMR data don’t provide the unambiguous assignment 09/10/2013 Protein Methods and Functions p. 33 of 69
Comparing NMR to X-ray n n n NMR family of structures often reflects real conformational heterogeneity Nonetheless, it’s hard to visualize what’s happening at the active site at any instant Hydrogens sometimes well-located in NMR; they’re often the least defined atoms in an Xray structure The NMR structure is obtained in solution! Hard to make NMR work if MW > 55 k. Da 09/10/2013 Protein Methods and Functions p. 34 of 69
What does it mean when NMR and X-ray structures differ? n n n Lattice forces may have tied down or moved surface amino acids in X-ray structure NMR may have errors in it X-ray may have errors in it (measurable) X-ray structure often closer to true atomic resolution X-ray structure has built-in reliability checks 09/10/2013 Protein Methods and Functions p. 35 of 69
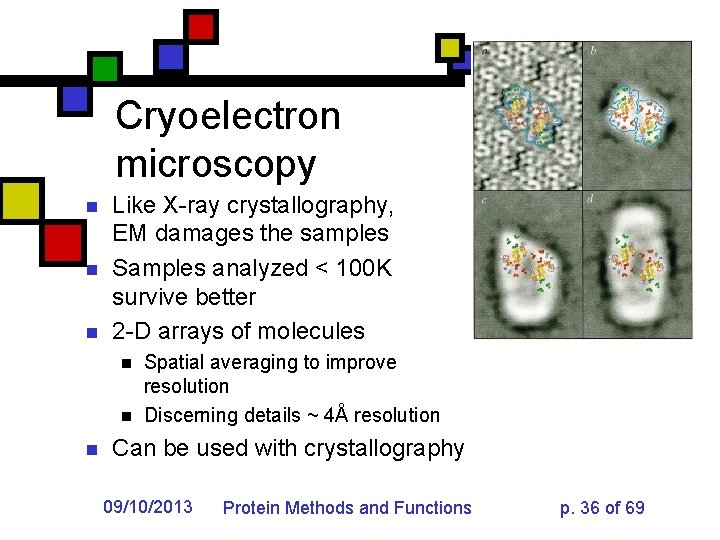
Cryoelectron microscopy n n n Like X-ray crystallography, EM damages the samples Samples analyzed < 100 K survive better 2 -D arrays of molecules n n n Spatial averaging to improve resolution Discerning details ~ 4Å resolution Can be used with crystallography 09/10/2013 Protein Methods and Functions p. 36 of 69
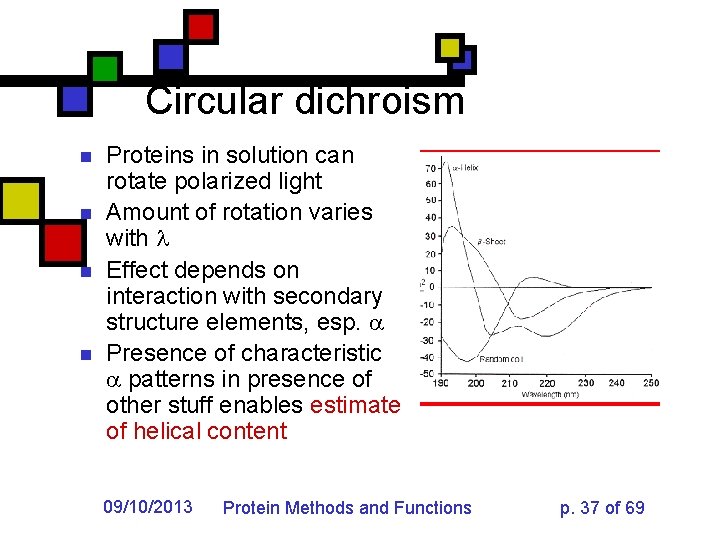
Circular dichroism n n Proteins in solution can rotate polarized light Amount of rotation varies with Effect depends on interaction with secondary structure elements, esp. Presence of characteristic patterns in presence of other stuff enables estimate of helical content 09/10/2013 Protein Methods and Functions p. 37 of 69
- Slides: 37