Protein Identification Using Mass Spectrometry Nathan Edwards Department















- Slides: 56

Protein Identification Using Mass Spectrometry Nathan Edwards Department of Biochemistry and Molecular & Cellular Biology Georgetown University Medical Center 12/8/2009 BIST 535 - 2009

Proteomics l Proteins are the machines that drive much of biology l l Genes are merely the recipe The direct characterization of a sample’s proteins en masse. l l 12/8/2009 What proteins are present? How much of each protein is present? BIST 535 - 2009 2

Systems Biology l Establish relationships by l l l Choosing related samples, Global characterization, and Comparison. Gene / Transcript / Protein Measurement Discrete (DNA) Continuous 12/8/2009 Predetermined Unknown Genotyping Sequencing Gene Expression Proteomics BIST 535 - 2009 3

Samples l l l Healthy / Diseased Cancerous / Benign Drug resistant / Drug susceptible Bound / Unbound Tissue specific Cellular location specific l 12/8/2009 Mitochondria, Membrane BIST 535 - 2009 4

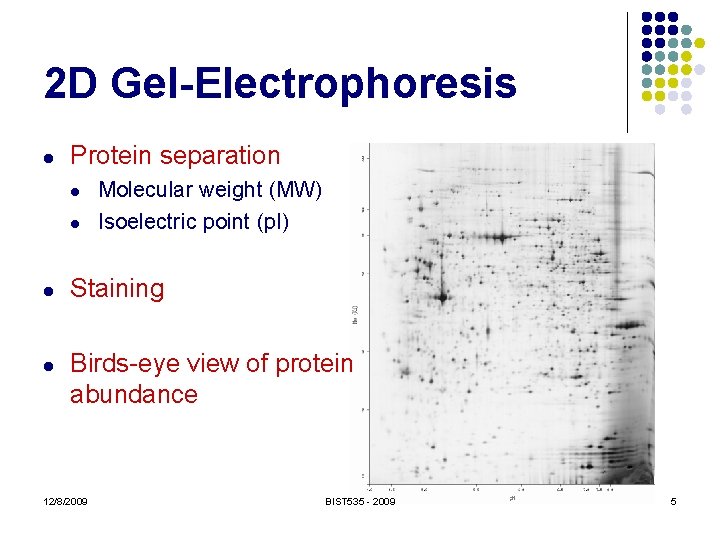
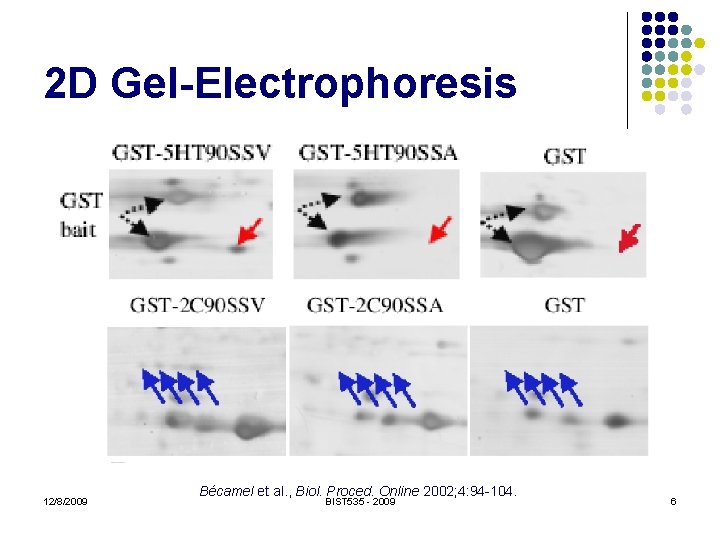
2 D Gel-Electrophoresis l Protein separation l l Molecular weight (MW) Isoelectric point (p. I) Staining Birds-eye view of protein abundance 12/8/2009 BIST 535 - 2009 5

2 D Gel-Electrophoresis 12/8/2009 Bécamel et al. , Biol. Proced. Online 2002; 4: 94 -104. BIST 535 - 2009 6

Paradigm Shift l l Traditional protein chemistry assay methods struggle to establish identity. Identity requires: l Specificity of measurement (Precision) l l A reference for comparison (Measurement → Identity) l 12/8/2009 Mass spectrometry Protein sequence databases BIST 535 - 2009 7

Mass Spectrometer Sample + _ Ionizer • MALDI • Electro-Spray Ionization (ESI) 12/8/2009 Mass Analyzer • Time-Of-Flight (TOF) • Quadrapole • Ion-Trap BIST 535 - 2009 Detector • Electron Multiplier (EM) 8

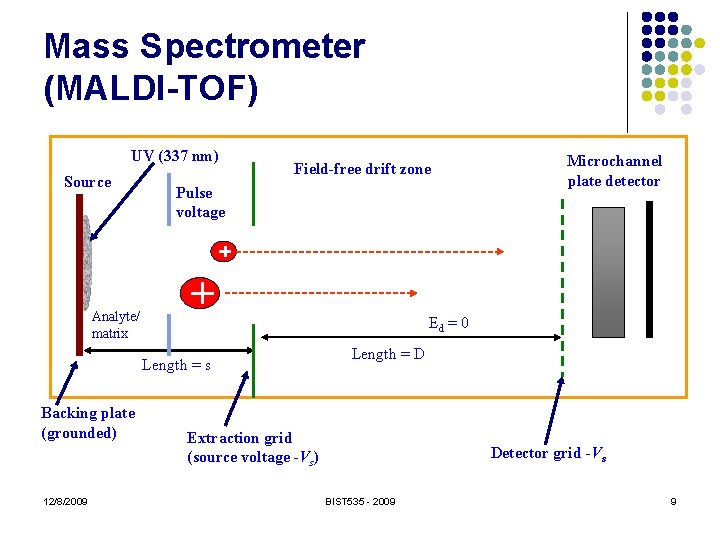
Mass Spectrometer (MALDI-TOF) UV (337 nm) Source Field-free drift zone Pulse voltage Analyte/ matrix Ed = 0 Length = s Backing plate (grounded) 12/8/2009 Microchannel plate detector Length = D Extraction grid (source voltage -Vs) Detector grid -Vs BIST 535 - 2009 9

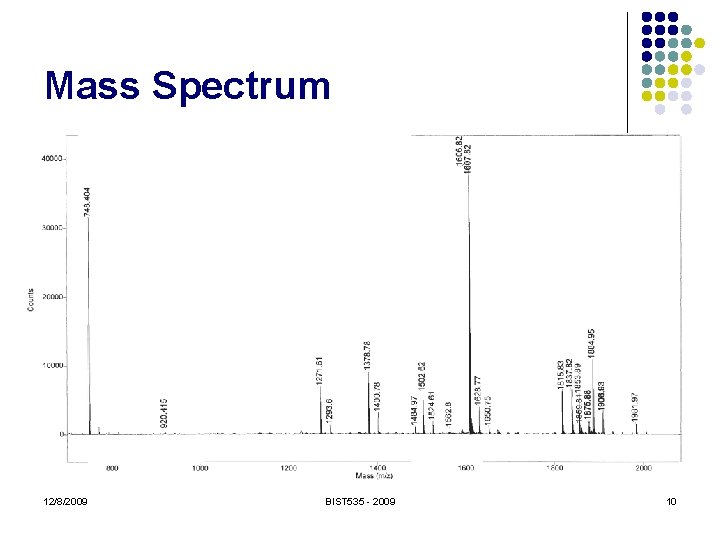
Mass Spectrum 12/8/2009 BIST 535 - 2009 10

Mass is fundamental 12/8/2009 BIST 535 - 2009 11

Sample Preparation for MS/MS Enzymatic Digest and Fractionation 12/8/2009 BIST 535 - 2009 12

Single Stage MS MS 12/8/2009 BIST 535 - 2009 13

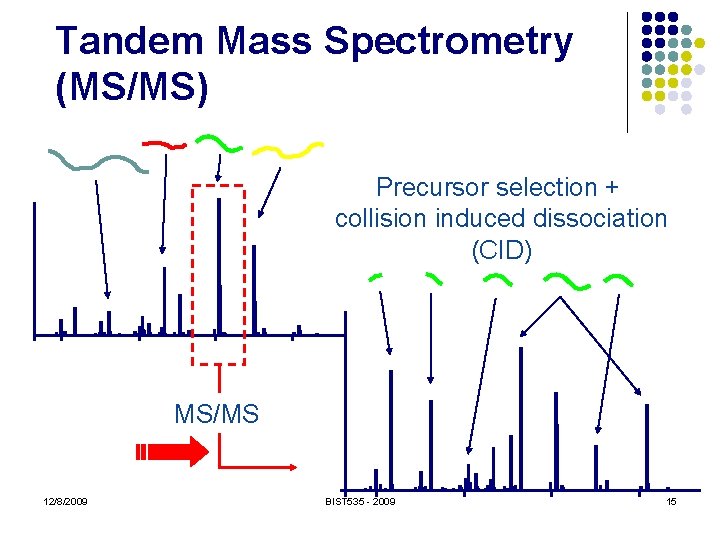
Tandem Mass Spectrometry (MS/MS) Precursor selection 12/8/2009 BIST 535 - 2009 14

Tandem Mass Spectrometry (MS/MS) Precursor selection + collision induced dissociation (CID) MS/MS 12/8/2009 BIST 535 - 2009 15

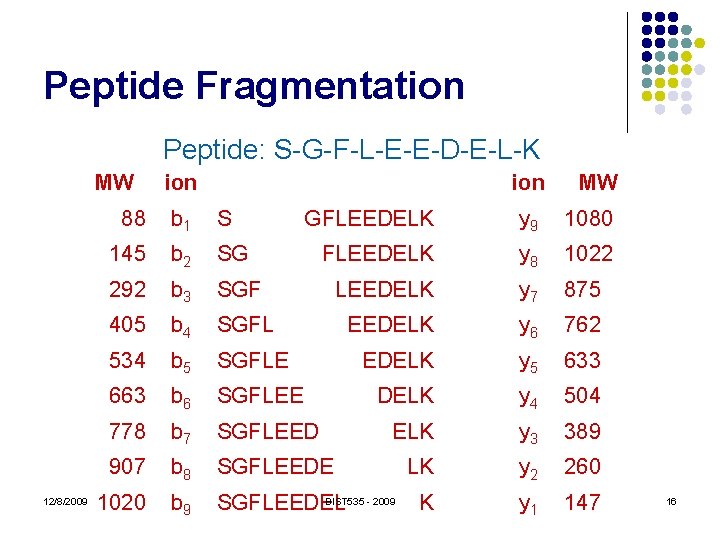
Peptide Fragmentation Peptide: S-G-F-L-E-E-D-E-L-K MW 12/8/2009 ion 88 b 1 S 145 b 2 SG 292 b 3 405 MW GFLEEDELK y 9 1080 FLEEDELK y 8 1022 SGF LEEDELK y 7 875 b 4 SGFL EEDELK y 6 762 534 b 5 SGFLE EDELK y 5 633 663 b 6 SGFLEE DELK y 4 504 778 b 7 SGFLEED ELK y 3 389 907 b 8 SGFLEEDE LK y 2 260 1020 b 9 BIST 535 - 2009 SGFLEEDEL K y 1 147 16

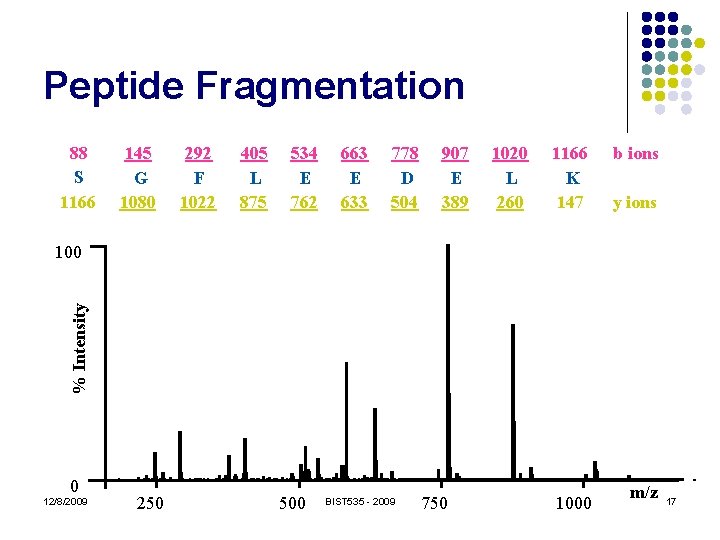
Peptide Fragmentation 88 S 1166 145 G 1080 292 F 1022 405 L 875 534 E 762 663 E 633 778 D 504 907 E 389 1020 L 260 1166 K 147 b ions y ions % Intensity 100 0 12/8/2009 250 500 BIST 535 - 2009 750 1000 m/z 17

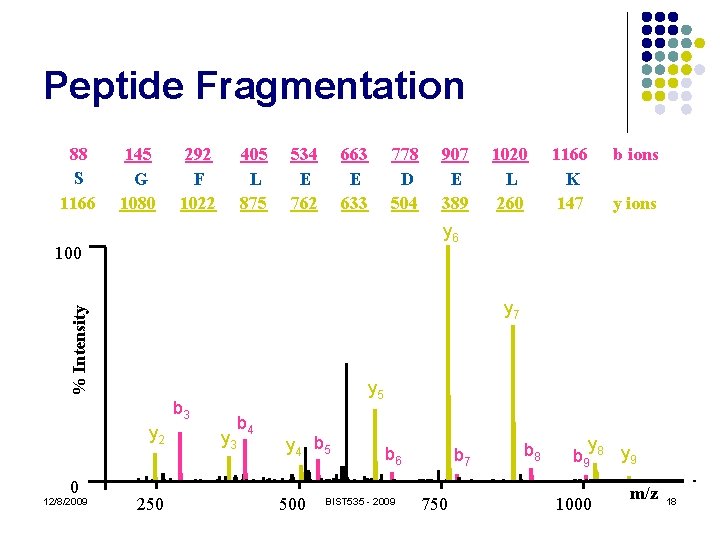
Peptide Fragmentation 88 S 1166 145 G 1080 292 F 1022 405 L 875 534 E 762 663 E 633 778 D 504 1166 K 147 b ions y ions % Intensity y 7 y 5 b 3 y 2 0 1020 L 260 y 6 100 12/8/2009 907 E 389 250 y 3 b 4 y 4 b 5 500 b 6 BIST 535 - 2009 b 7 750 b 8 y b 9 8 y 9 1000 m/z 18

Peptide Identification Given: l The mass of the precursor ion, and l The MS/MS spectrum Output: l The amino-acid sequence of the peptide 12/8/2009 BIST 535 - 2009 19

Sequence Database Search l l Compares peptides from a protein sequence database with spectra Filter peptide candidates by l l l Precursor mass Digest motif Score each peptide against spectrum l l l 12/8/2009 Generate all possible peptide fragments Match putative fragments with peaks Score and rank BIST 535 - 2009 20

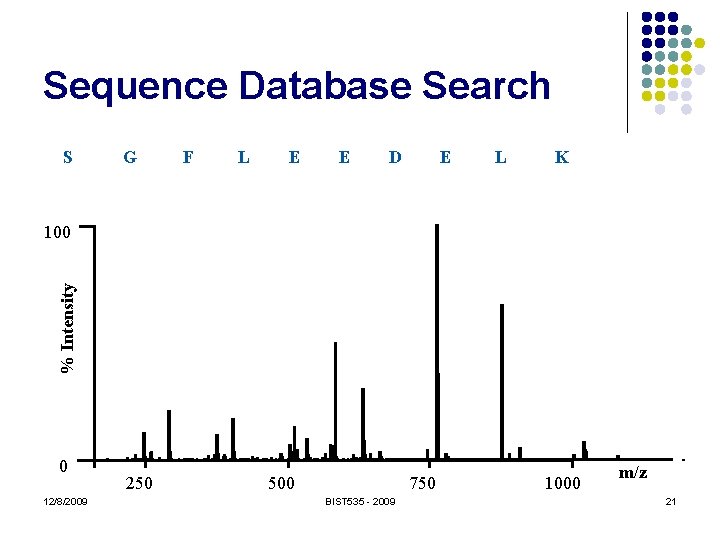
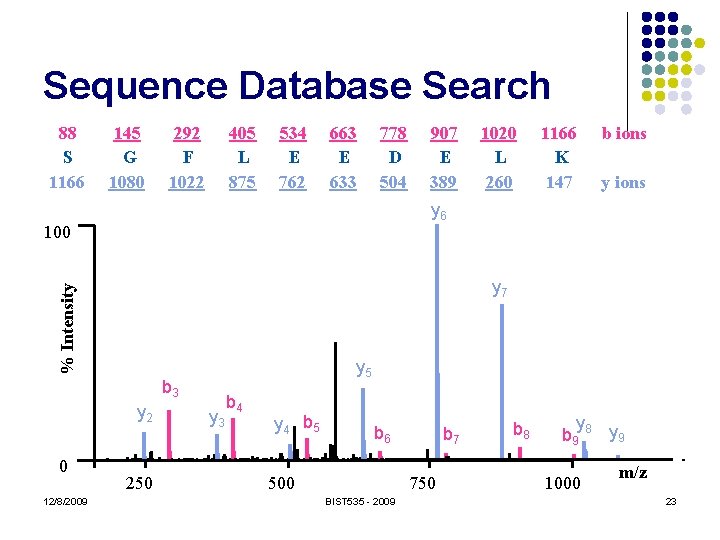
Sequence Database Search S G F L E E D E L K % Intensity 100 0 12/8/2009 250 500 750 BIST 535 - 2009 1000 m/z 21

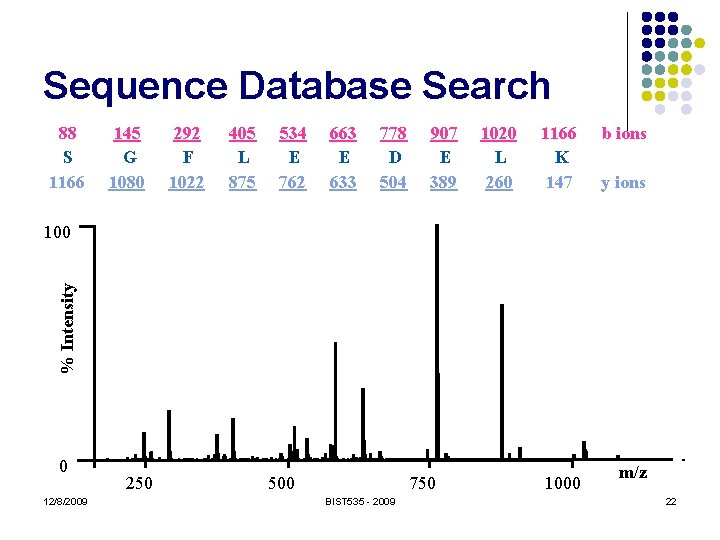
Sequence Database Search 88 S 1166 145 G 1080 292 F 1022 405 L 875 534 E 762 663 E 633 778 D 504 907 E 389 1020 L 260 1166 K 147 b ions y ions % Intensity 100 0 12/8/2009 250 500 750 BIST 535 - 2009 1000 m/z 22

Sequence Database Search 88 S 1166 145 G 1080 292 F 1022 405 L 875 534 E 762 663 E 633 778 D 504 1166 K 147 b ions y ions % Intensity y 7 y 5 b 3 y 2 12/8/2009 1020 L 260 y 6 100 0 907 E 389 250 y 3 b 4 y 4 b 5 b 6 500 b 7 750 BIST 535 - 2009 b 8 y b 9 8 y 9 1000 m/z 23

Sequence Database Search l No need for complete ladders l Possible to model all known peptide fragments l Sequence permutations eliminated l All candidates have some biological relevance l Practical for high-throughput peptide identification l Correct peptide might be missing from database! 12/8/2009 BIST 535 - 2009 24

Peptide Candidate Filtering l l l Digestion Enzyme: Trypsin Cuts just after K or R unless followed by a P. Basic residues (K & R) at C-terminal attract ionizing charge, leading to strong y-ions “Average” peptide length about 10 -15 aminoacids Must allow for “missed” cleavage sites 12/8/2009 BIST 535 - 2009 25

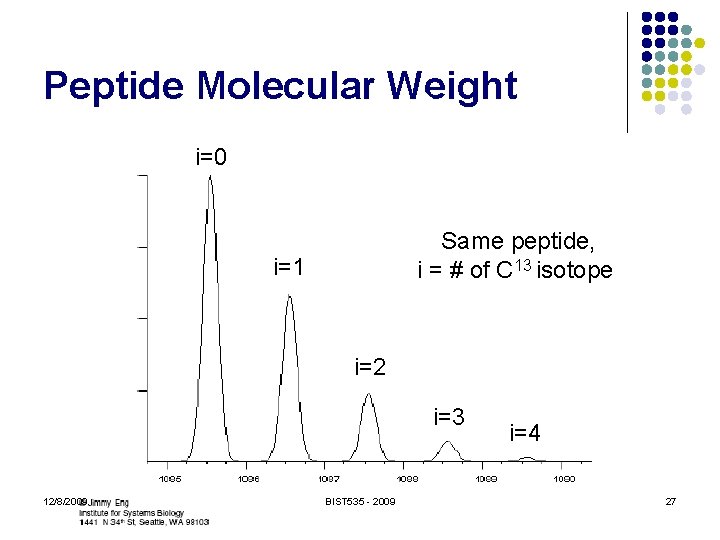
Peptide Candidate Filtering l l Peptide molecular weight Only have m/z value l l l Need to determine charge state Ion selection tolerance Mass for each amino-acid symbol? l l 12/8/2009 Monoisotopic vs. Average “Default” residue mass Depends on sample preparation protocol Cysteine almost always modified BIST 535 - 2009 26

Peptide Molecular Weight i=0 Same peptide, i = # of C 13 isotope i=1 i=2 i=3 12/8/2009 BIST 535 - 2009 i=4 27

Peptide Scoring l Peptide fragments vary based on l l l The instrument The peptide’s amino-acid sequence The peptide’s charge state Etc… Search engines model peptide fragmentation to various degrees. l l 12/8/2009 Speed vs. sensitivity tradeoff y-ions & b-ions occur most frequently BIST 535 - 2009 28

Peptide Identification l l High-throughput workflows demand we analyze all spectra, all the time. Spectra may not contain enough information to be interpreted correctly l l Peptides may not match our assumptions l l …bad static on a cell phone …its all Greek to me “Don’t know” is an acceptable answer! 12/8/2009 BIST 535 - 2009 29

Peptide Identification l Rank the best peptide identifications l Is the top ranked peptide correct? 12/8/2009 BIST 535 - 2009 30

Peptide Identification l Rank the best peptide identifications l Is the top ranked peptide correct? 12/8/2009 BIST 535 - 2009 31

Peptide Identification l Rank the best peptide identifications l Is the top ranked peptide correct? 12/8/2009 BIST 535 - 2009 32

Peptide Identification l Incorrect peptide has best score l l Correct peptide is missing? Potential for incorrect conclusion What score ensures no incorrect peptides? Correct peptide has weak score l l l 12/8/2009 Insufficient fragmentation, poor score Potential for weakened conclusion What score ensures we find all correct peptides? BIST 535 - 2009 33

Statistical Significance l Can’t prove particular identifications are right or wrong. . . l l A minimal standard for identification scores. . . l l l . . . need to know fragmentation in advance! . . . better than guessing. p-value, E-value, statistical significance For each spectrum, compare scores with those of random peptides (p-value, E-value). 12/8/2009 BIST 535 - 2009 34

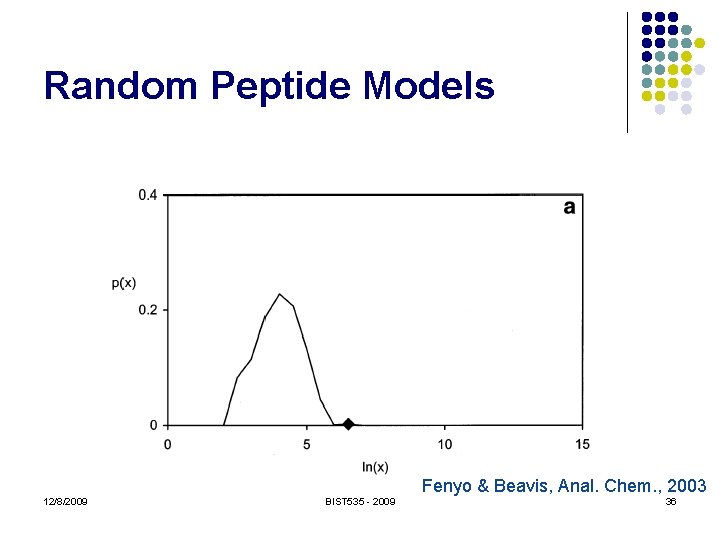
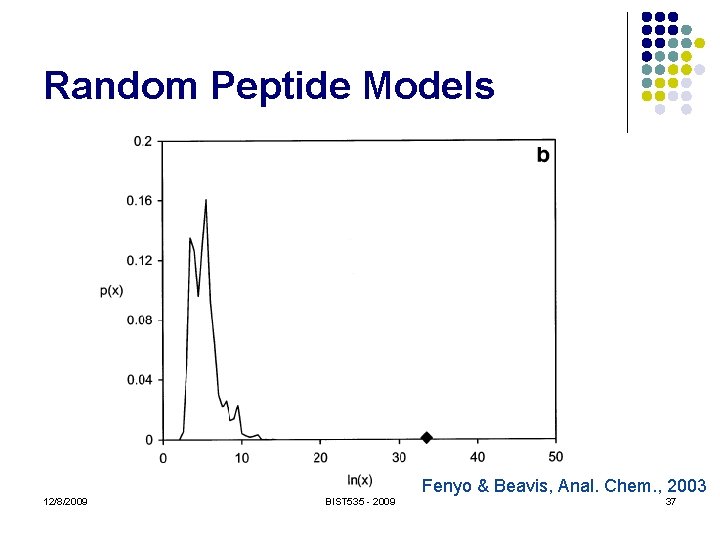
Random Peptide Models l "Generate" random peptides l l l Real looking fragment masses No theoretical model! Must use empirical distribution Usually require they have the correct precursor mass Score function can model anything we like! 12/8/2009 BIST 535 - 2009 35

Random Peptide Models Fenyo & Beavis, Anal. Chem. , 2003 12/8/2009 BIST 535 - 2009 36

Random Peptide Models Fenyo & Beavis, Anal. Chem. , 2003 12/8/2009 BIST 535 - 2009 37

Random Peptide Models l l l Truly random peptides don’t look much like real peptides Just use (incorrect) peptides from the sequence database! Caveats: l l l Correct peptide (non-random) may be included Peptides are not independent Reverse sequence avoids only the first problem 12/8/2009 BIST 535 - 2009 38

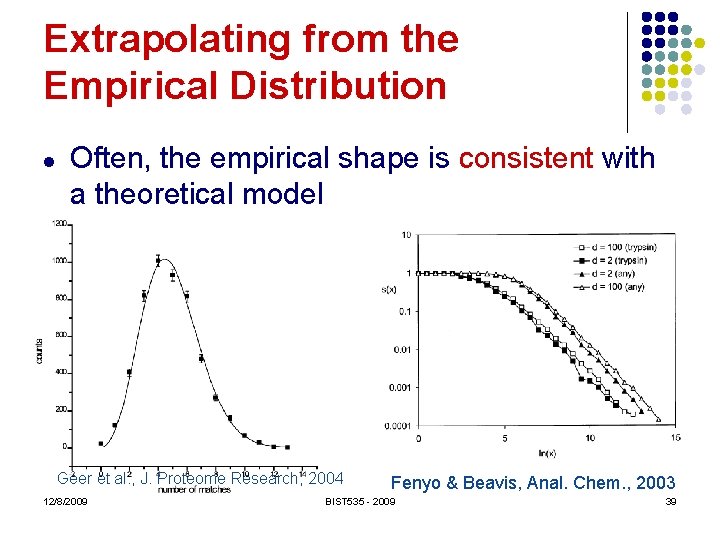
Extrapolating from the Empirical Distribution l Often, the empirical shape is consistent with a theoretical model Geer et al. , J. Proteome Research, 2004 12/8/2009 Fenyo & Beavis, Anal. Chem. , 2003 BIST 535 - 2009 39

False Positive Rate Estimation l Each spectrum is a chance to be right, wrong, or inconclusive. l l l Given identification criteria: l l l At any given threshold, how many peptide identifications are wrong? Computed for entire spectral dataset SEQUEST Xcorr, E-value, Score, etc. , plus. . . threshold Use “decoy” sequences and repeat search l l 12/8/2009 random, reverse, cross-species Identifications must be incorrect! BIST 535 - 2009 40

False Positive Rate Estimation l # FP in real search = # hits in decoy search l l Need same size database, or rate conversion FP Rate: # decoy hits with score ≥ thresh # hits with score ≥ thresh 12/8/2009 BIST 535 - 2009 41

False Positive Rate Estimation l A form of statistical significance l Search engine independent l l Easy to implement Assumes a single threshold for all spectra l 12/8/2009 Best if E-value or similar is used to compute a spectrum normalized score BIST 535 - 2009 42

Peptide Prophet l From the Institute for Systems Biology l l Re-analysis of SEQUEST results l l Keller et al. , Anal. Chem. 2002 Spectrum dependant scores (XCorr) Assumes that many of the spectra are not correctly identified 12/8/2009 BIST 535 - 2009 43

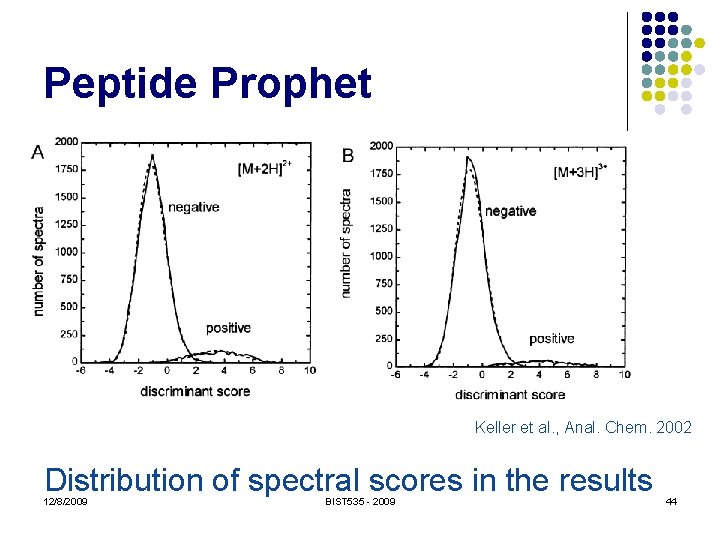
Peptide Prophet Keller et al. , Anal. Chem. 2002 Distribution of spectral scores in the results 12/8/2009 BIST 535 - 2009 44

Peptide Prophet l l Assumes a bimodal distribution of scores, with a particular shape Ignores database size l l …but it is included implicitly Like empirical distribution for peptide sampling, can be applied to any score function l 12/8/2009 Can be applied to any search engines’ results BIST 535 - 2009 45

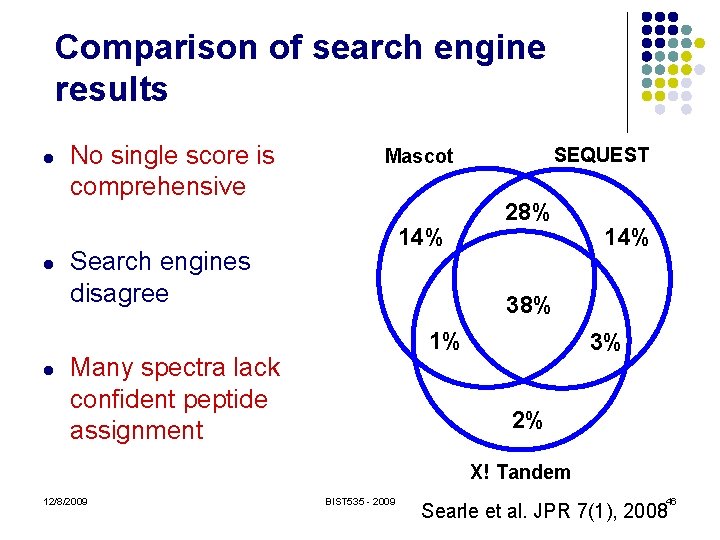
Comparison of search engine results l l l No single score is comprehensive SEQUEST Mascot 14% Search engines disagree 28% 14% 38% 1% Many spectra lack confident peptide assignment 3% 2% X! Tandem 12/8/2009 BIST 535 - 2009 46 Searle et al. JPR 7(1), 2008

Combining search engine results – harder than it looks! l Consensus boosts confidence, but. . . l l How to handle weak identifications? l l l How to assess statistical significance? Gain specificity, but lose sensitivity! Incorrect identifications are correlated too! Consensus vs disagreement vs abstention Threshold at some significance? We apply unsupervised machine-learning. . l 12/8/2009 Lots of related work unified in a single framework. BIST 535 - 2009 47

Supervised Learning 12/8/2009 BIST 535 - 2009 48

Unsupervised Learning 12/8/2009 BIST 535 - 2009 49

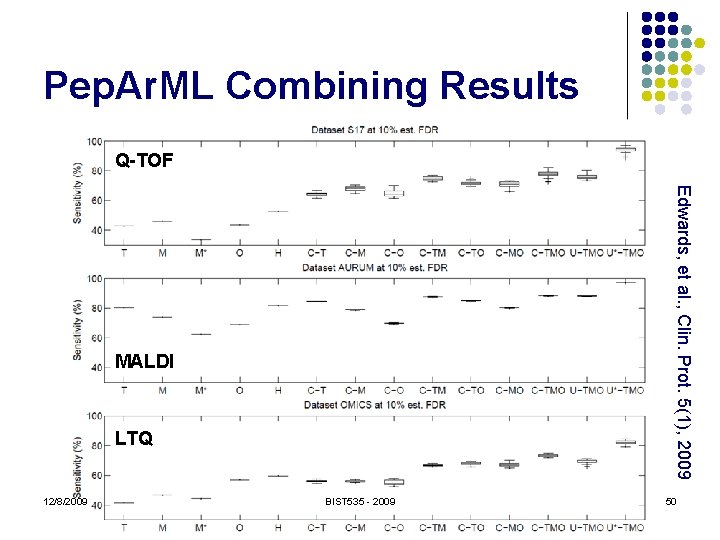
Pep. Ar. ML Combining Results Q-TOF Edwards, et al. , Clin. Prot. 5(1), 2009 MALDI LTQ 12/8/2009 BIST 535 - 2009 50

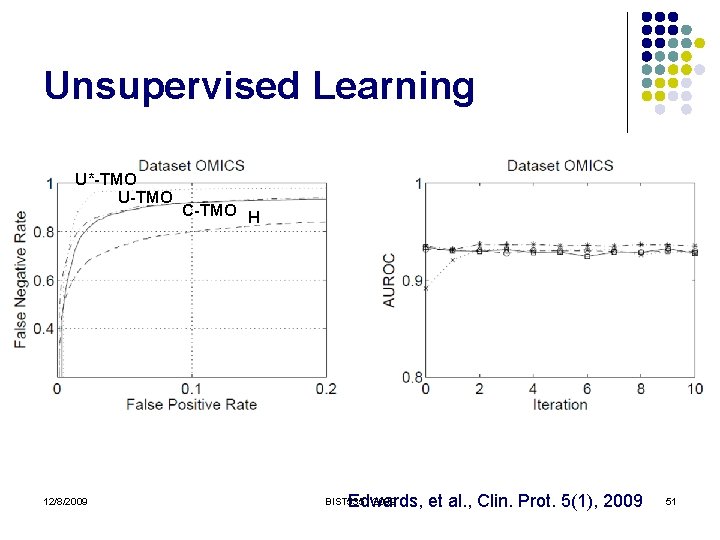
Unsupervised Learning U*-TMO U-TMO 12/8/2009 C-TMO H Edwards, et al. , Clin. Prot. 5(1), 2009 BIST 535 - 2009 51

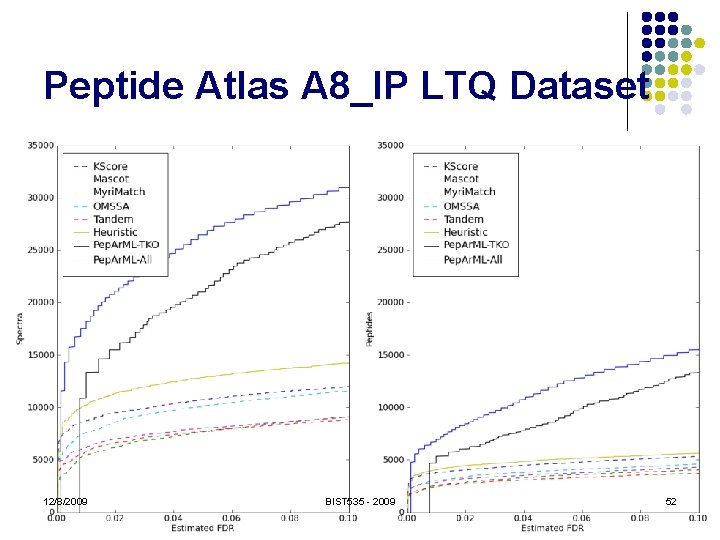
Peptide Atlas A 8_IP LTQ Dataset 12/8/2009 BIST 535 - 2009 52

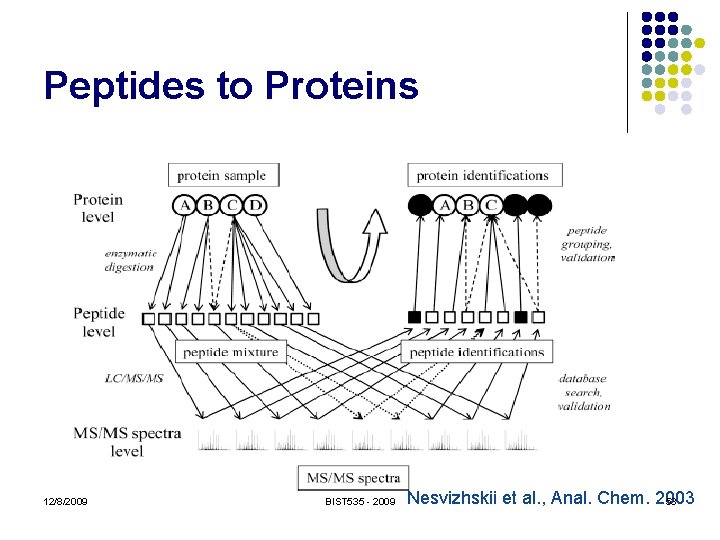
Peptides to Proteins 12/8/2009 BIST 535 - 2009 Nesvizhskii et al. , Anal. Chem. 2003 53

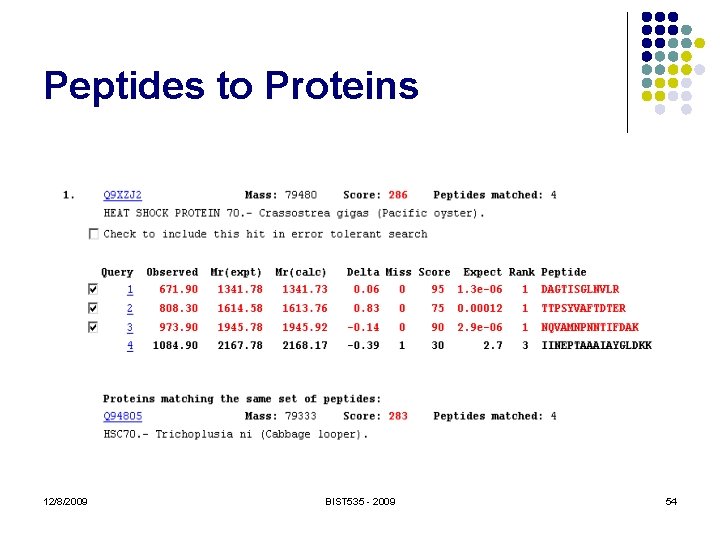
Peptides to Proteins 12/8/2009 BIST 535 - 2009 54

Peptides to Proteins l A peptide sequence may occur in many different protein sequences l l Variants, paralogues, protein families Separation, digestion and ionization is not well understood Proteins in sequence database are extremely non-random, and very dependent No great tools for assessing statistical confidence of protein identifications. 12/8/2009 BIST 535 - 2009 55

Summary l l Protein identification from tandem mass spectra is a key proteomics technology. Protein identifications should be treated with healthy skepticism. l l All peptide / protein lists represent a triage of the data – look for ways to estimate significance. Lots of open "applied statistics" problems! l 12/8/2009 The devil is in the details – there is no highmoral ground here – whatever is most effective wins. BIST 535 - 2009 56