Protein functions prediction Swiss Institute of Bioinformatics Institut
Protein functions prediction Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Introduction n n n Signal peptides Transmembrane regions and topology PTM (post-translational modifications) Low complexity and biased regions Repeats Coils n n n Secondary structure Antigenic peptides Domain/Motifs Tools The EMBOSS package Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Different techniques n Algorithms n n n Sliding window, Nearest Neighbor Patterns, regular expression Weight matrices HMM, profiles Neural Networks Rules Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
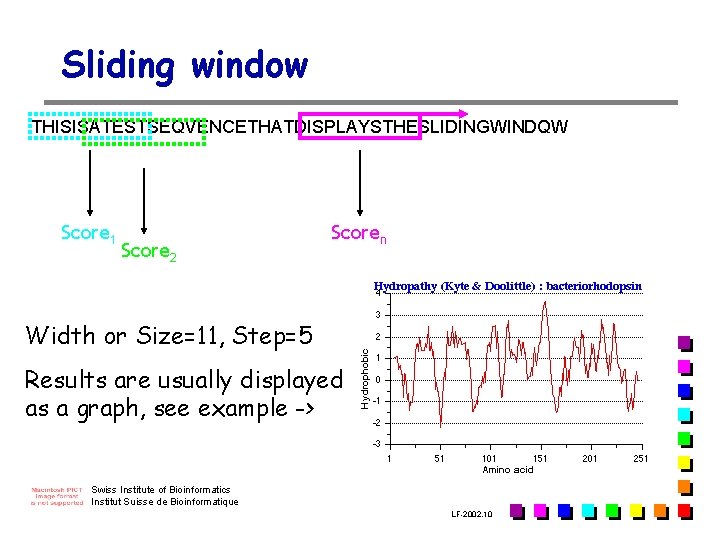
Sliding window THISISATESTSEQVENCETHATDISPLAYSTHESLIDINGWINDQW Score 1 Score 2 Scoren Width or Size=11, Step=5 Results are usually displayed as a graph, see example -> Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
-x(0, 1)-{V} Regexp: ^A. [ST]{2}. ? [^V] Patterns / regular expression n n Pattern: <A-x-[ST](2)-x(0, 1)-{V} Regexp: ^A. [ST]{2}. ? [^V]](http://slidetodoc.com/presentation_image_h/12996d9bc8e99fbe0a3e67a23fba8a05/image-5.jpg)
Patterns / regular expression n n Pattern: <A-x-[ST](2)-x(0, 1)-{V} Regexp: ^A. [ST]{2}. ? [^V] Text: The sequence must start with an alanine, followed by any amino acid, followed by a serine or a threonine, two times, followed by any amino acid or nothing, followed by any amino acid except a valine. Simply the syntax differ… Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
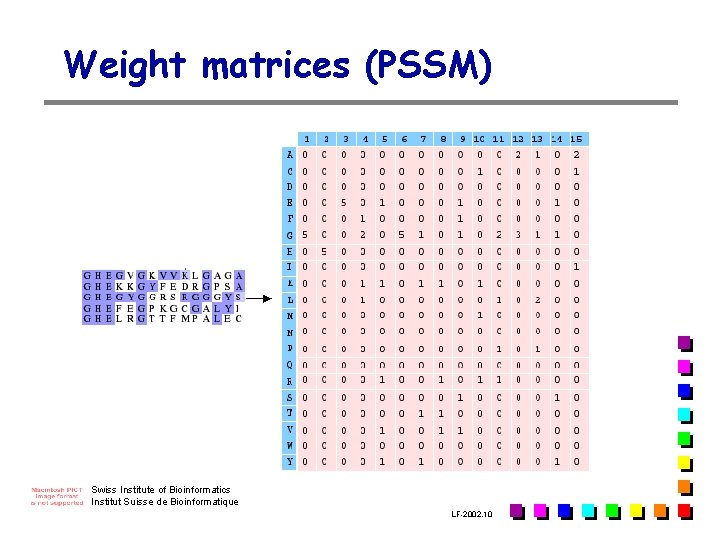
Weight matrices (PSSM) Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
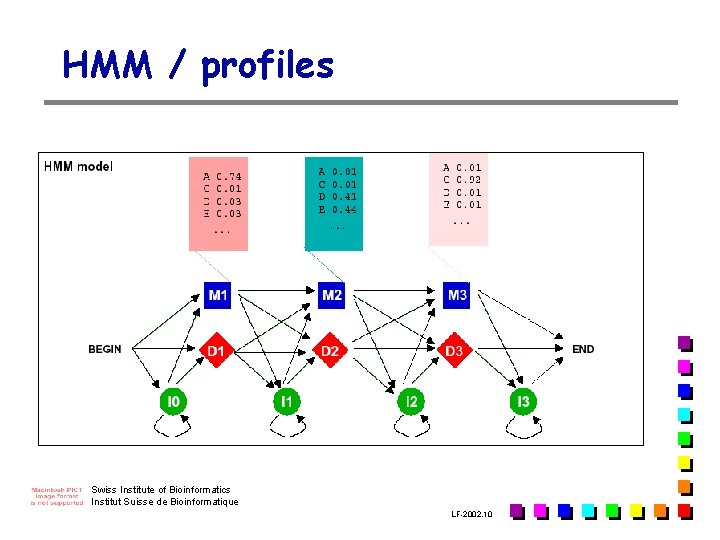
HMM / profiles Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
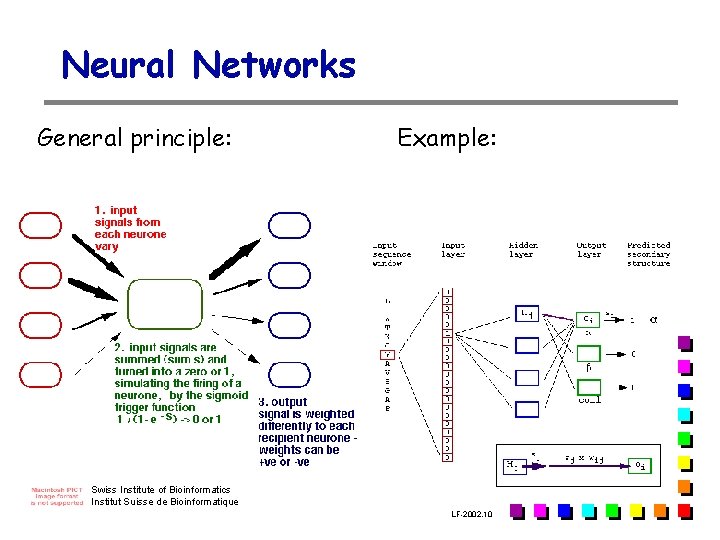
Neural Networks General principle: Example: Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Signals found in proteins n N-ter n n exportation - secretion mitochondria chloroplast n n n internal n NLS (nuclear localization signal) C-ter n GPI-anchor (Glycosyl Phosphatidyl Inositol) other membrane anchors (see PTM) other unknown ? Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Signals detection tools n n n n Signal. P Mito. Prot Chloro. P Predotar PSort Target. P Sigcleave (EMBOSS) n n Big-PI DGPI Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
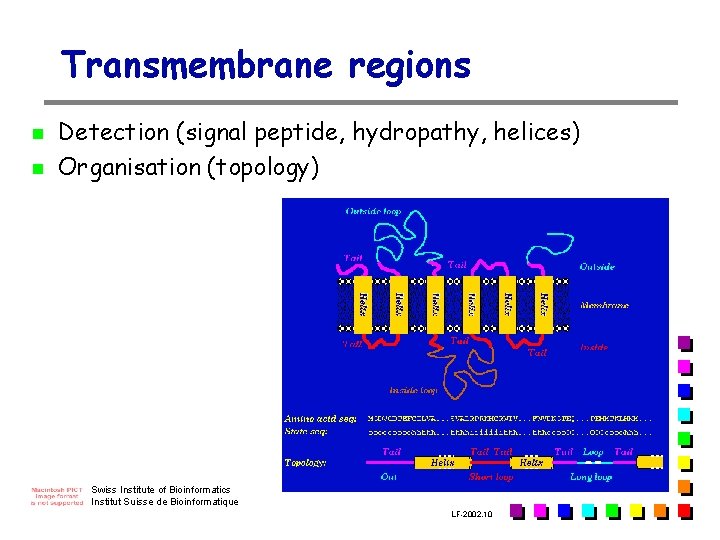
Transmembrane regions n n Detection (signal peptide, hydropathy, helices) Organisation (topology) Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Transmembrane detection tools n n n TMHMM TMPred Top. Pred 2 DAS HMMTop Tmap (EMBOSS) Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Post translational modifications n Phosphorylation n n S - T - (HO)K Acetylation, methylation n n N O-glycosylation n n S-T-Y N-glycosylation n D-E-K Sulfation n Y Farnesylation, myristylation, palmitoylation, geranylation, GPIanchor n n Ubiquitination and family n n n C - Nter - Cter K - Nter Inteins (protein splicing) Pre-translational n Selenoprotein n C Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
PTM detection n n Pattern prediction (PROSITE) Short or weak signal Frequent hit producer Best method is experimental n n n MS/MS detection Most method use « rules » joining pattern detection and knowledge to predict sites. n n n Net. OGlyc - Prediction of type Oglycosylation sites in mammalian proteins Dicty. OGlyc - Prediction of Glc. NAc O-glycosylation sites in Dictyostelium Yin. OYang - O-beta-Glc. NAc attachment sites in eukaryotic protein sequences Net. Phos - Prediction of Ser, Thr and Tyr phosphorylation sites in eukaryotic proteins NMT - Prediction of N-terminal Nmyristoylation Sulfinator - Prediction of tyrosine sulfation sites Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Low complexity regions n n n repeats compositional bias PEST Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Low complexity / Repeats n DUST (DNA) / SEG n n n search collection REPRO, Radar n n REP n de novo detection EMBOSS (DNA) n Repeat. Masker (DNA) n n de novo detection n einverted equicktandem etandem palindrome EMBOSS (protein) n oddcomp PEST, PESTFind n de novo detection Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
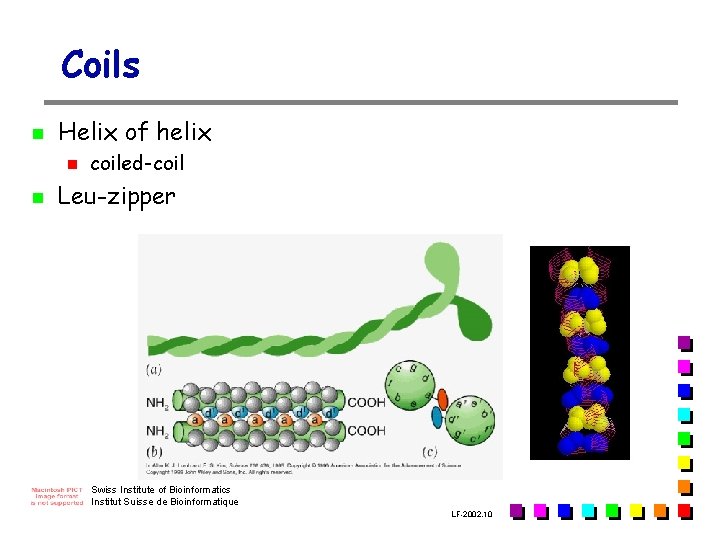
Coils n Helix of helix n n coiled-coil Leu-zipper Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Coils detection n COILS n n Paircoil, Multicoil n n Pairwise correlation Marcoil n n Weight matrices HMM Pepcoil (EMBOSS) n Weight matrices Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Secondary structure n Structure to predict n n Alpha-helices Beta-sheets Turns Random coil n n n n Garnier (EMBOSS) PHD DSC PREDATOR NNSSP Jpred Jnet Many others Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Antigenic peptide n n Peptides binding to MHC class I n n n 15 mers (3+9+3) Depend highly on MHC type Use of experimental knowledge n 8, 9, 10 mers class II n n n n Databases of known peptides SYFPEITHI HLA_Bind (BIMAS) MAPPP combined expert Antigenic (EMBOSS) Many more n Prediction of proteasome cleavage sites n n Net. Chop Pa. Proc Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
Domain / Motif n All the protein domain descriptors n n n n PROSITE PFAM SMART PRODOM BLOCKS PRINTS … n n Federation: Inter. Pro Many techniques n n Patterns, Regexp PSSM (PSI-BLAST) Profiles HMM Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10

Other Tools n You can find some of them on our servers n n Or on Ex. PASy server n n www. ch. embnet. org www. expasy. org/tools Or ask Google!! n www. google. com Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
European Molecular Biology Open Software Suite Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10

n n Free Open Source (for most Unix plateforms) GCG successor (compatible with GCG file format) More than 100 programs Easy to install locally n n n Interfaces n n n but no interface, requires local databases Unix command-line only Jemboss, www 2 gcg, w 2 h, wemboss … (with account) Pise, EMBOSS-GUI (no account) Access: http: //www. emboss. org Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
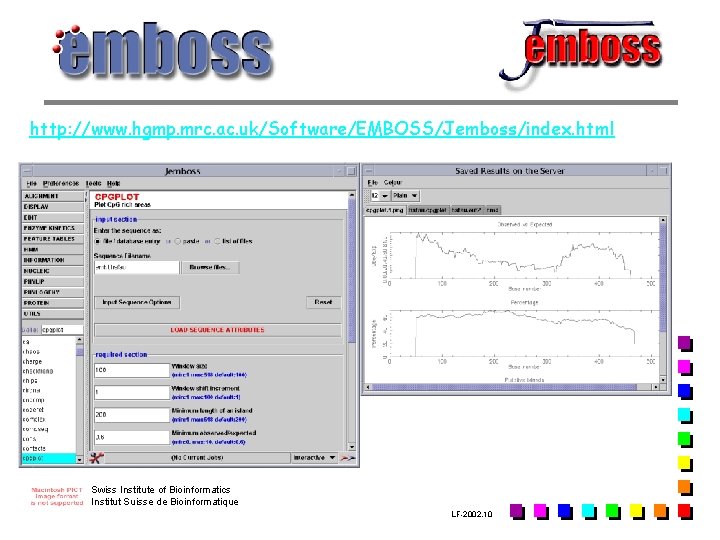
http: //www. hgmp. mrc. ac. uk/Software/EMBOSS/Jemboss/index. html Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
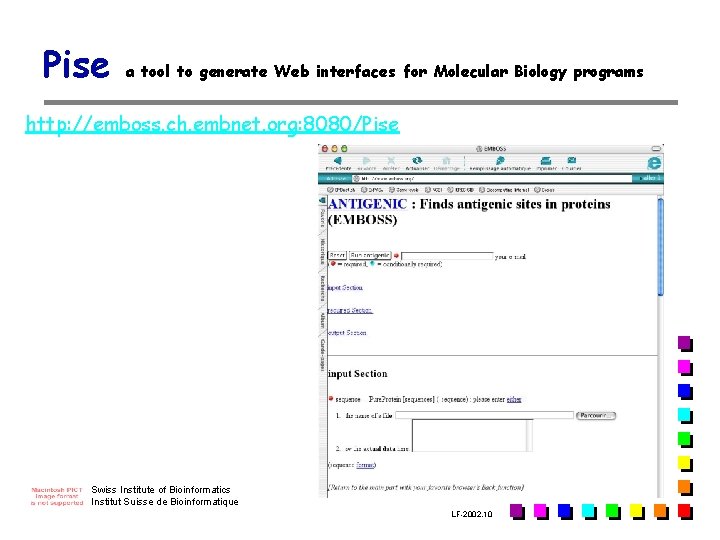
Pise a tool to generate Web interfaces for Molecular Biology programs http: //emboss. ch. embnet. org: 8080/Pise Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
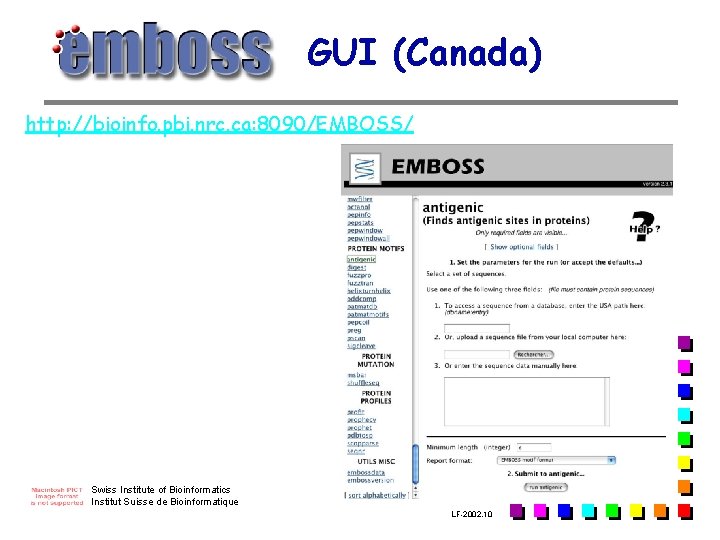
GUI (Canada) http: //bioinfo. pbi. nrc. ca: 8090/EMBOSS/ Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
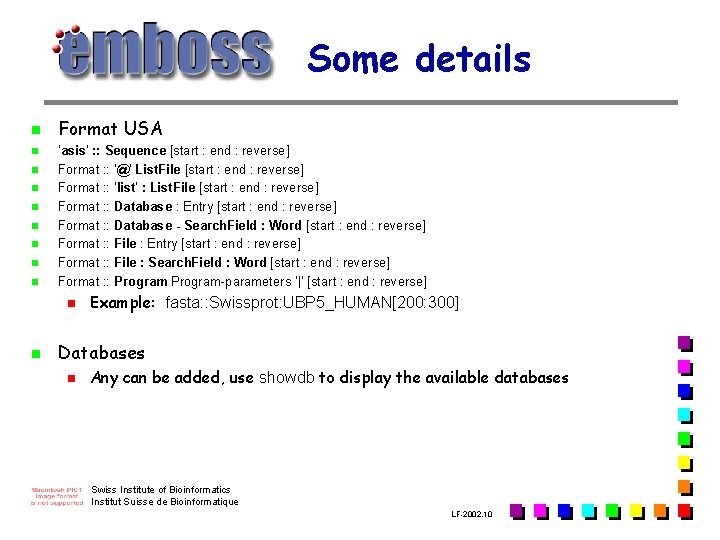
Some details n n n n n Format USA 'asis' : : Sequence [start : end : reverse] Format : : '@' List. File [start : end : reverse] Format : : 'list' : List. File [start : end : reverse] Format : : Database : Entry [start : end : reverse] Format : : Database - Search. Field : Word [start : end : reverse] Format : : File : Entry [start : end : reverse] Format : : File : Search. Field : Word [start : end : reverse] Format : : Program-parameters '|' [start : end : reverse] n n Example: fasta: : Swissprot: UBP 5_HUMAN[200: 300] Databases n Any can be added, use showdb to display the available databases Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
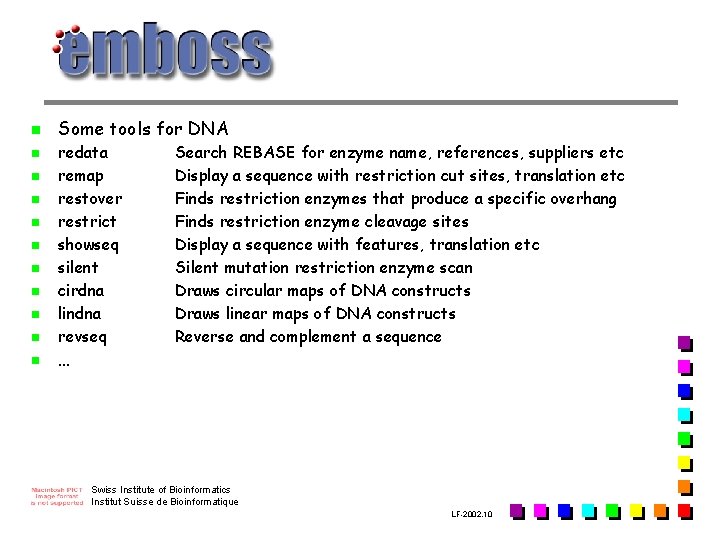
n n n Some tools for DNA redata remap restover restrict showseq silent cirdna lindna revseq … Search REBASE for enzyme name, references, suppliers etc Display a sequence with restriction cut sites, translation etc Finds restriction enzymes that produce a specific overhang Finds restriction enzyme cleavage sites Display a sequence with features, translation etc Silent mutation restriction enzyme scan Draws circular maps of DNA constructs Draws linear maps of DNA constructs Reverse and complement a sequence Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
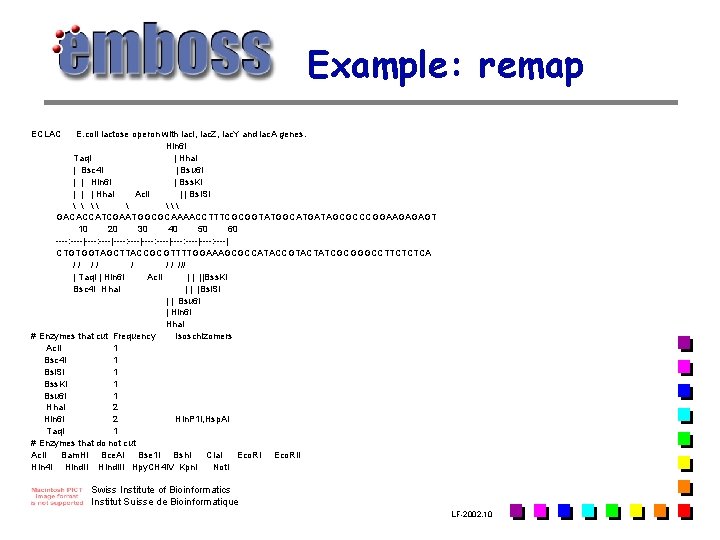
Example: remap ECLAC E. coli lactose operon with lac. I, lac. Z, lac. Y and lac. A genes. Hin 6 I Taq. I | Hha. I | Bsc 4 I | Bsu 6 I | | Hin 6 I | Bss. KI | | | Hha. I Aci. I | | Bsi. SI \ GACACCATCGAATGGCGCAAAACCTTTCGCGGTATGGCATGATAGCGCCCGGAAGAGAGT 10 20 30 40 50 60 ----: ----|----: ----| CTGTGGTAGCTTACCGCGTTTTGGAAAGCGCCATACCGTACTATCGCGGGCCTTCTCTCA // /// | Taq. I | Hin 6 I Aci. I | | ||Bss. KI Bsc 4 I Hha. I | | |Bsi. SI | | Bsu 6 I | Hin 6 I Hha. I # Enzymes that cut Frequency Isoschizomers Aci. I 1 Bsc 4 I 1 Bsi. SI 1 Bss. KI 1 Bsu 6 I 1 Hha. I 2 Hin 6 I 2 Hin. P 1 I, Hsp. AI Taq. I 1 # Enzymes that do not cut Acl. I Bam. HI Bce. AI Bse 1 I Bsh. I Cla. I Eco. RII Hin 4 I Hind. III Hpy. CH 4 IV Kpn. I Not. I Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
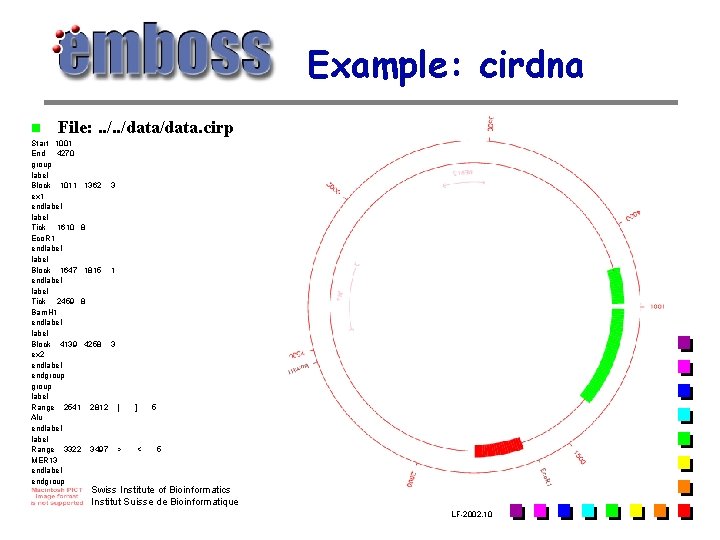
Example: cirdna n File: . . /data/data. cirp Start 1001 End 4270 group label Block 1011 1362 3 ex 1 endlabel Tick 1610 8 Eco. R 1 endlabel Block 1647 1815 1 endlabel Tick 2459 8 Bam. H 1 endlabel Block 4139 4258 3 ex 2 endlabel endgroup label Range 2541 2812 [ Alu endlabel Range 3322 3497 > MER 13 endlabel endgroup ] 5 < 5 Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10

Exercises n DEA Exercises web based sequence analysis n The goal of this exercise is to use web based tools for protein sequence analysis n n n c) use the different profile, motifs, pattern databases to get more information about the domain(s) you found. d) How do you evaluate the PRINTS tropomyosin annotation in this Tr. EMBL entry (Q 9 WZH 0)? List of useful links: n n n a) Take this Tr. EMBL sequence (Q 9 X 252) and try a BLAST against swissprot with the complete protein or with the first 70 residues. Explain the difference. Use TMPred, Signal. P, and COILS to help you. b) Pass this sequence through PFSCAN and search all databases. Compare with this command on ludwig -sun 1/2: hits -b "prf pat pfam" tr: Q 9 X 252 basic BLAST or advanced BLAST or PSI-BLAST TMPred prediction tool for transmembrane regions (or TMHMM) COILS prediction tool for coiled-coil regions Signal. P prediction tool for signal-peptide cleavage site Profile, domain, motifs databases and search sites: n n n PFSCAN Inter. Pro (Pfam, PRINTS, PROSITE, SMART) HITS Swiss Institute of Bioinformatics Institut Suisse de Bioinformatique LF-2002. 10
- Slides: 32