Protein Folding Processing and Degradation Protein Folding Protein

























- Slides: 52

Protein Folding, Processing and Degradation

Protein Folding • Protein in native state is not static – 2° structural elements and domains move • Function of proteins often dependent on large conformational changes, triggered by ligand binding 2

Energy of Folding • Difference in energy (free energy) between folded (native) and unfolded (denatured) state is small, 5 -15 kcal/mol. • Two major contributions to energy difference between unfolded and folded state, enthalpy and entropy 3

Free energy of folding • Enthalpy (H) is increased upon folding – Noncovalent interactions are maximized • Stronger and more frequent in native state – Enthalpy difference can reach several hundred kcal/mol • Entropy is decreased upon folding – Folding causes one main conformation=highly ordered structure – Entropy difference can reach several hundred kcal/mol 4

Free energy of folding • Enthalpy and entropy differences balance each • • other, and DG is a small positive number. Small DG is necessary because too large a free energy change would mean a very stable protein, one that would never change However, structural flexibility is important to protein function, and proteins need to be degraded 5

Protein folding • For any given protein, there is one conformation • • that has significantly lower free energy than any other state Achieved through kinetic pathway of unstable intermediates (not all intermediates are sampled) Assisted by chaperones and protein disulfide isomerases so intermediates are not trapped in a local low energy state 6

Pathways to folding • There are multiple folding pathways for some proteins, but a single main pathway for other proteins 7

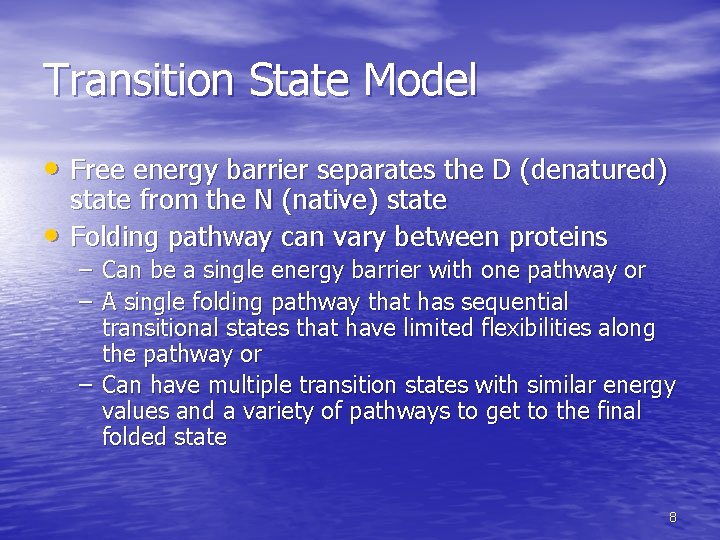
Transition State Model • Free energy barrier separates the D (denatured) • state from the N (native) state Folding pathway can vary between proteins – Can be a single energy barrier with one pathway or – A single folding pathway that has sequential transitional states that have limited flexibilities along the pathway or – Can have multiple transition states with similar energy values and a variety of pathways to get to the final folded state 8

Fig. 6 -39, 9 p. 189

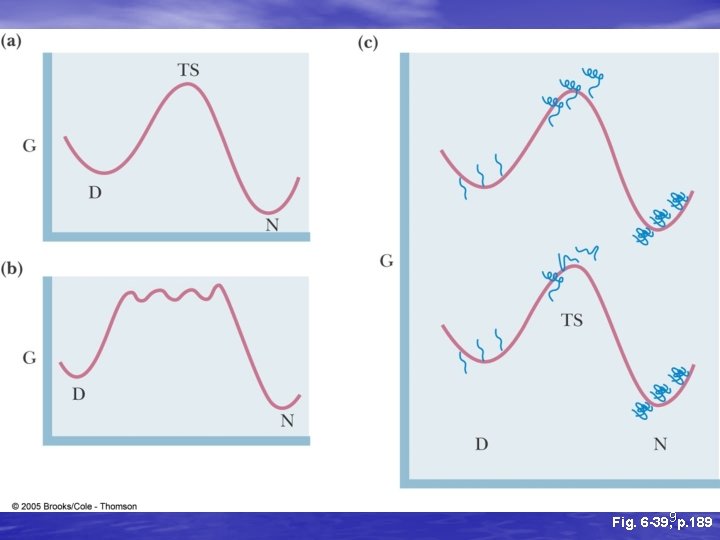
10 p. 190 Fig. 6 -40,

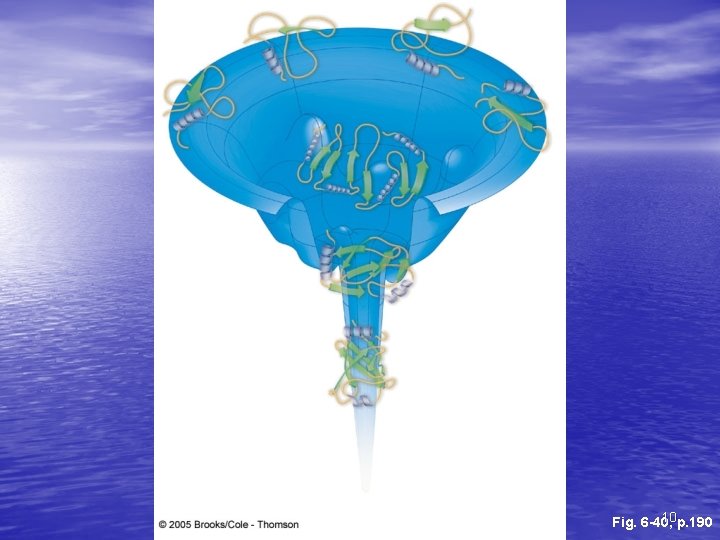
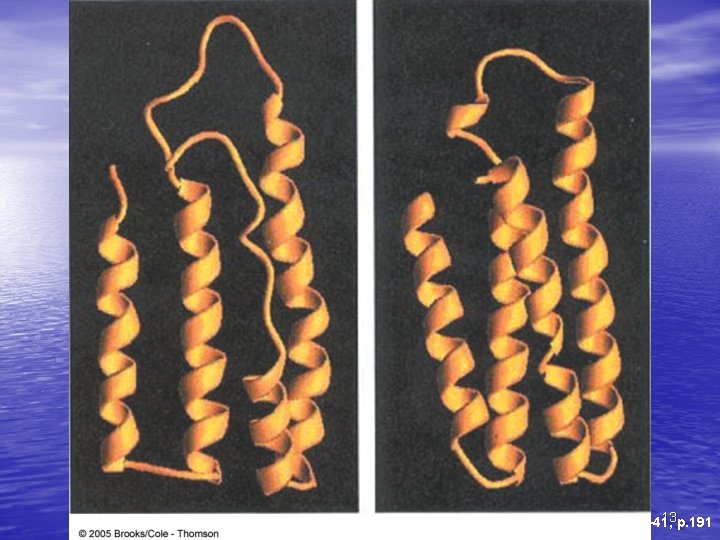
Pathways to folded state • Molten globular is state in between unfolded and final folded state – Can be multiple molten globular states for each protein • Has helices and sheets, which may be positioned • correctly Unfolded loops, less compaction, different conformations of surface structures 11

fast Denatured slow Molten globular Native 12

13 p. 191 Fig. 6 -41,

Disulfide bond formation • Cellular environment is reducing • Cysteines must be oxidized to form S-S bonds • Bacteria use Disulfide bridge-forming protein, Dsb, for oxidation in periplasm • In eucaryotes, protein disulfide isomerase, PDI, used for oxidation in the endoplasmic reticulum (ER) 14

Dsb • Crystal structure shows a thioredoxin-like domain • Mechanism of thioredoxin is based on reversible • oxidation of two cysteine thiol groups to a disulfide-redox-active disulfide bridge is Cys-X-X-Cys (X any amino acid) Oxidized Dsb is less stable than reduced form-– S-S is very reactive, therefore a strong oxidizing agent 15

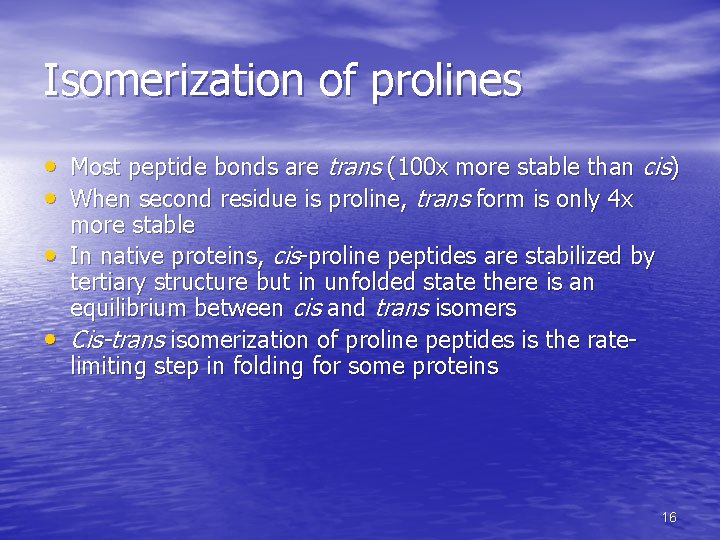
Isomerization of prolines • Most peptide bonds are trans (100 x more stable than cis) • When second residue is proline, trans form is only 4 x • • more stable In native proteins, cis-proline peptides are stabilized by tertiary structure but in unfolded state there is an equilibrium between cis and trans isomers Cis-trans isomerization of proline peptides is the ratelimiting step in folding for some proteins 16

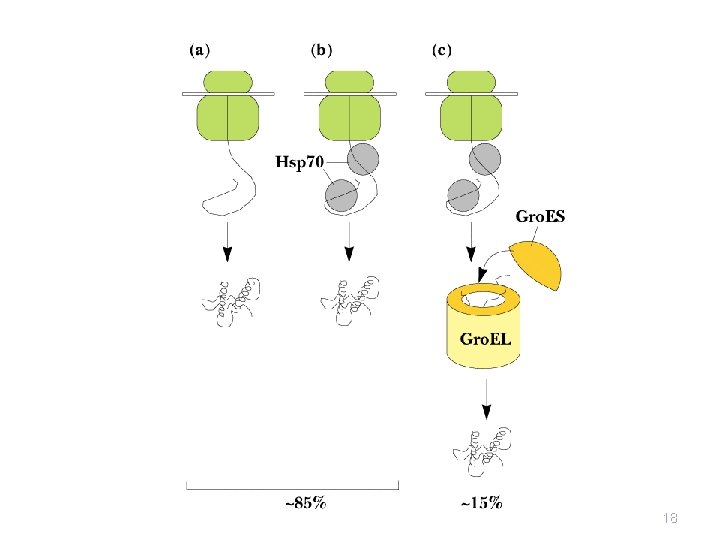
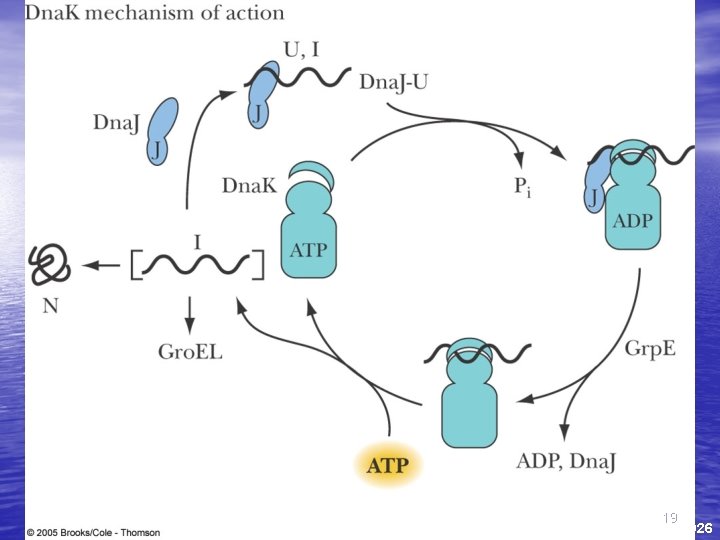
Protein Folding • Proteins are assisted in folding by molecular chaperones • Hsp 60 (chaperonins) Hsp 70 and Hsp 90 are three main classes • Hsp 70 recognizes exposed, unfolded regions of new protein chains - especially hydrophobic regions • It binds to these regions, protecting them until productive folding reactions can occur 17

18

19 Fig. 31 -2 b, p. 1026

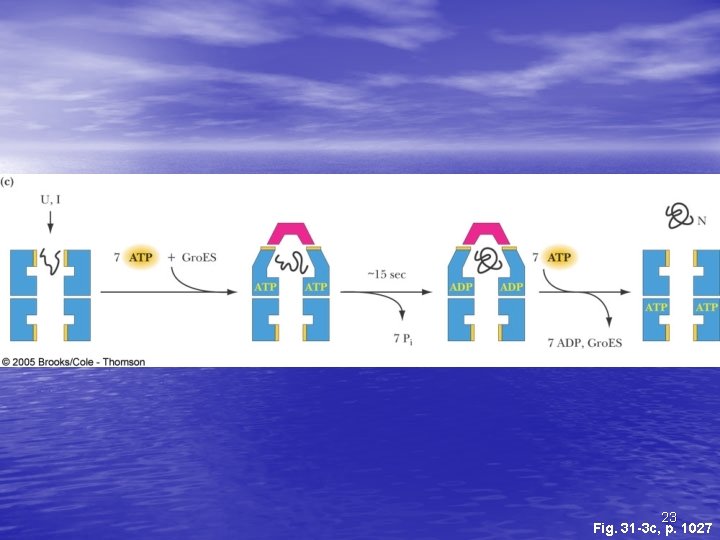
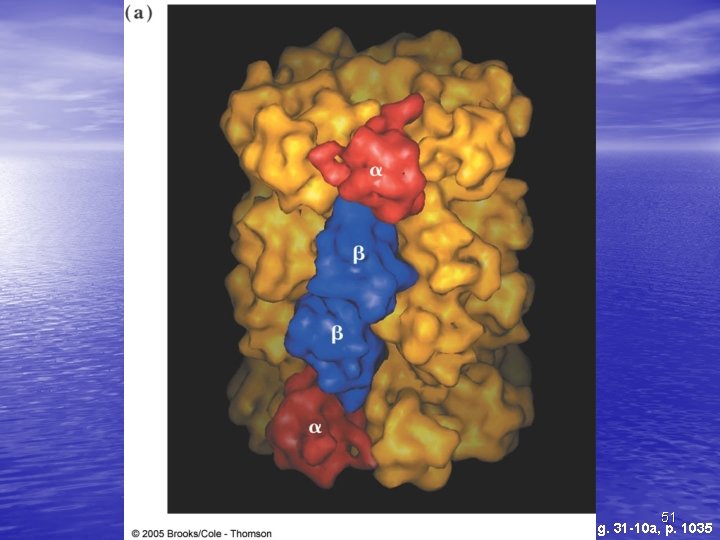
The Gro. ES-Gro. EL Complex • The principal chaperonin in E. coli • Gro. EL forms two stacked 7 -membered rings of • • 60 k. D subunits; Gro. ES is a dome on the top Nascent protein apparently binds reversibly many times to the walls of the donut structure, each time driven by ATP hydrolysis, eventually adopting its folded structure, then being released from the Gro. ES-Gro. EL complex Rhodanese (as one example) requires hydrolysis of 130 ATP to reach fully folded state 20

21 Fig. 31 -3 a, p. 1027

22 Fig. 31 -3 b, p. 1027

23 Fig. 31 -3 c, p. 1027

Eucaryotic Hsp 90 Chaperones • Hsp 90 is 1 -2% of total cytosolic protein • Signal transduction molecules such as tyrosine kinase receptors, steroid hormone receptors, non-receptor tyrosine kinases are clients for Hsp 90 24

Fate of nascent proteins • Association of nascent polypeptide with various chaperone systems commits them to folding pathways • If proteins fail to fold, they are recognized and targeted for degradation 25

Protein processing • Post-translational modifications are covalent modifications that alter function • Hundreds of amino acid variations have been described • Methylation, phosphorylation, lipid and carbohydrate addition are a few • Human proteins number around 300, 000 from 30, 000 genes 26

Proteolytic cleavage • Why remove amino acids? – Create diversity • Met-amidopeptidase removes Met from peptide so that not all amino termini are Met – Serves as activation mechanism • Metabolically active enzymes are pro-proteins or zymogens such as digestive enzymes – Involves targeting of proteins to proper destinations 27

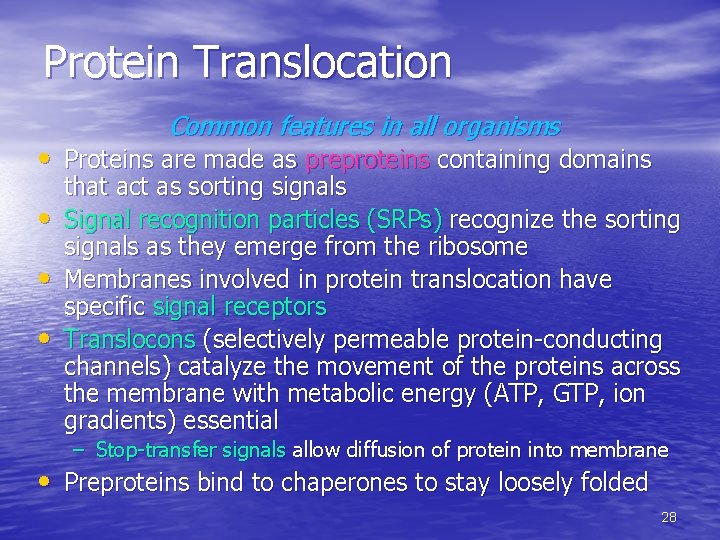
Protein Translocation Common features in all organisms • Proteins are made as preproteins containing domains • • • that act as sorting signals Signal recognition particles (SRPs) recognize the sorting signals as they emerge from the ribosome Membranes involved in protein translocation have specific signal receptors Translocons (selectively permeable protein-conducting channels) catalyze the movement of the proteins across the membrane with metabolic energy (ATP, GTP, ion gradients) essential – Stop-transfer signals allow diffusion of protein into membrane • Preproteins bind to chaperones to stay loosely folded 28

Prokaryotic Protein Translocation An essential process for membrane proteins and secretory proteins • Such proteins are synthesized with a leader • • peptide, or signal sequence of about 16 -26 amino acids The signal sequence has a basic N-terminus, a central domain of 7 -13 hydrophobic residues, and a nonhelical C-terminus The signal sequence directs the newly synthesized protein to its proper destination 29

Prokaryotic Protein Transport All non-cytoplasmic proteins must be translocated • The leader peptide retards the folding of the protein so that molecular chaperone proteins can interact with it and direct its folding • The leader peptide also provides recognition signals for the translocation machinery • A leader peptidase removes the leader sequence when folding and targeting are assured 30

Eukaryotic Protein Sorting Eukaryotic cells contain many membrane-bounded compartments • Most (but not all) targeting sequences are N • • terminal, cleaveable presequences Charge distribution, polarity and secondary structure of the signal sequence, rather than a particular sequence, appears to target to particular organelles and membranes Synthesis of secretory and membrane proteins is coupled to translocation across ER membrane 31

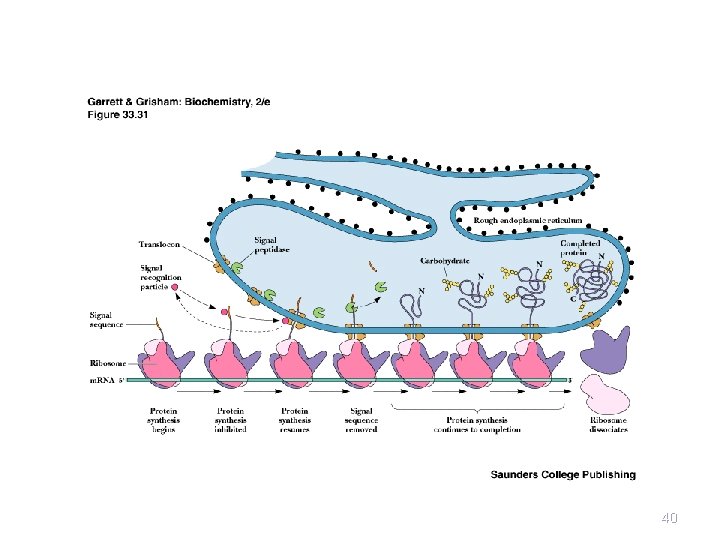
Events at the ER Membrane • As the signal sequence emerges from the ribosome, a signal recognition particle (SRP) finds it and escorts it to the ER membrane • There it docks with a docking protein or signal receptor (SR) - see Figure 31. 5 • SRP dissociates in a GTP-dependent process • Protein synthesis resumes and protein passes into ER or into ER membrane; signal is cleaved 32

33

34

35

36

37

Ribosome dissociates 38

39

40

Signal Recognition Particle • Complex of six polypepides and one 300 - base RNA molecule • Signal peptide recognition depends on one protein of 54, 000 daltons-SRP 54 • RNA (7 S) is probably structural • Removal of RNA disrupts SRP 41

Mitochondrial protein import • Mitochondria have two membranes, and two spaces in between the membranes • Signal sequences are N-terminal, positively charged regions of 10 -70 aa • Form amphipathic a-helices, positive on one side and uncharged, hydrophobic on the other (Fig. 31. 6) 42

Mitochondrial protein import (Fig 31. 7) • Binding of preprotein to TOM (mitochondrial outer • • • membrane translocon) For outer membrane proteins, transfer to SAM (sorting and assembly complex) Inner membrane proteins traverse TOM and are taken up by TIM 22 (inner mitochondrial membrane translocon) Matrix proteins are taken up by TIM 23 and transferred across the inner membrane into the matrix 43

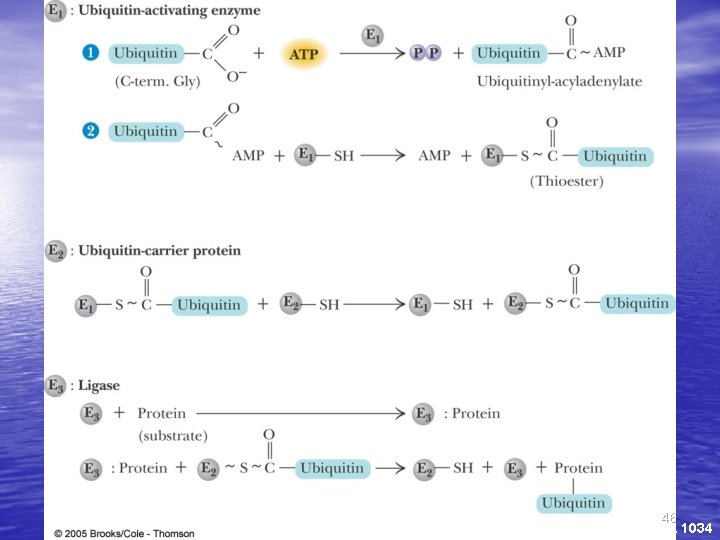
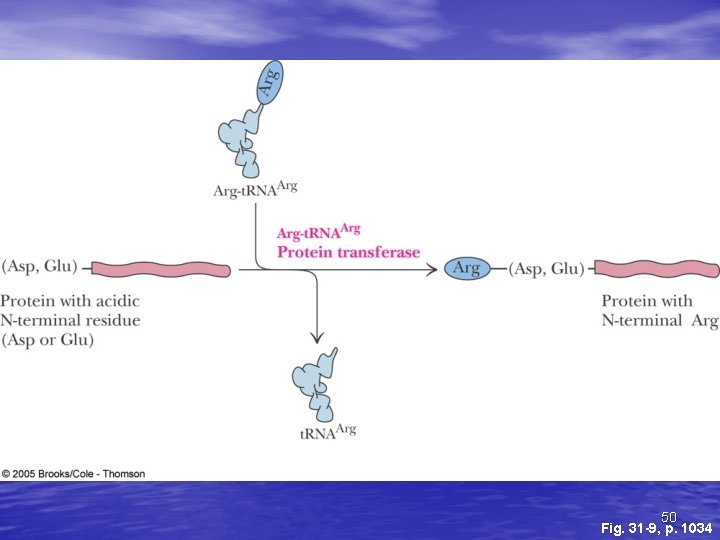
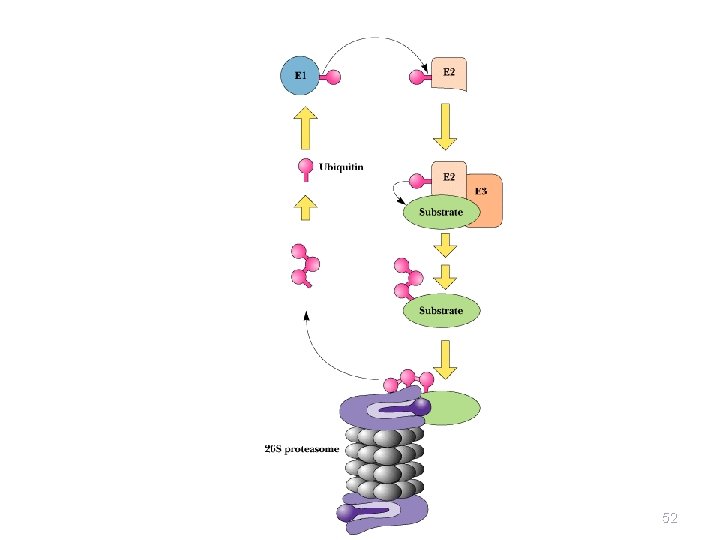
Protein Degradation • Some protein degradation pathways are • • • nonspecific - randomly cleaved proteins seem to be rapidly degraded However, there is also a selective, ATPdependent pathway for degradation - the ubiquitin-mediated pathway Ubiquitin is a highly-conserved, 76 residue (8. 5 k. D) protein found widely in eukaryotes Proteins are committed to degradation by conjugation with ubiquitin 44

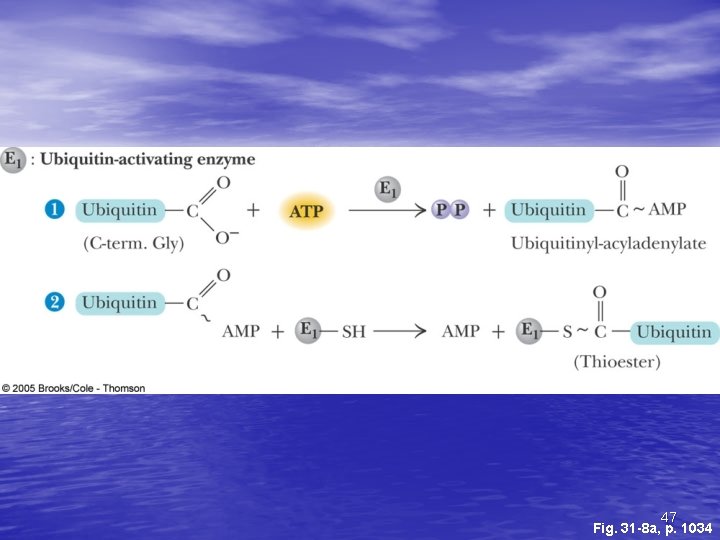
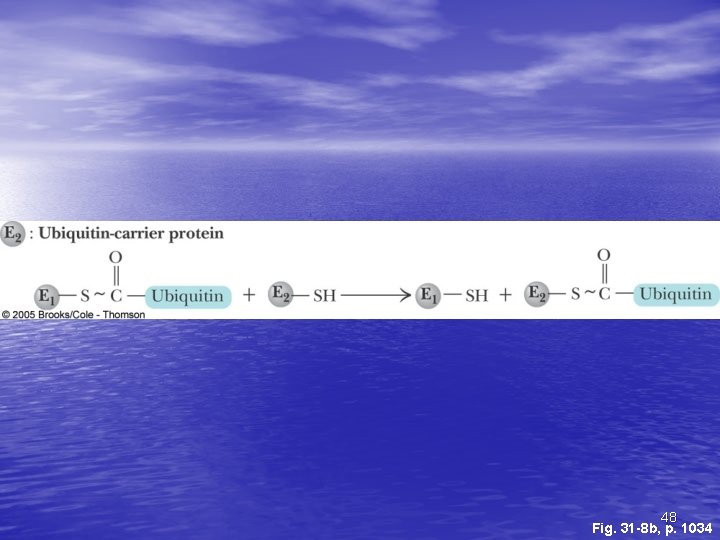
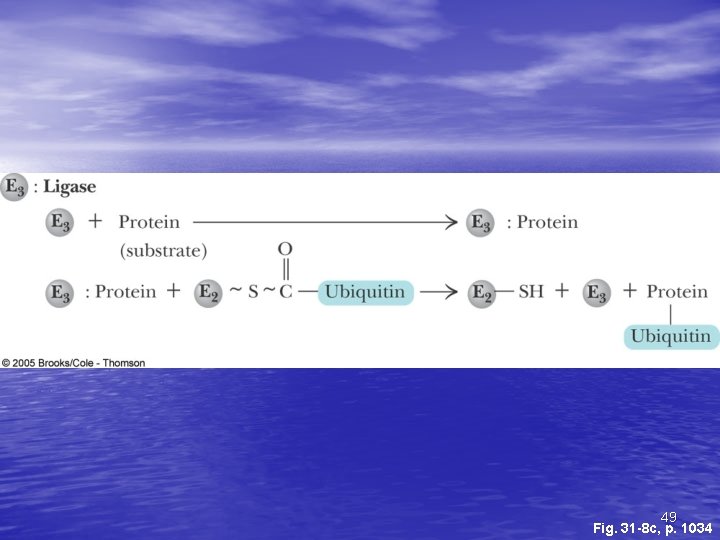
Ubiquitin and Degradation Three proteins involved: E 1, E 2 and E 3 • E 1 is the ubiquitin-activating enzyme - it forms a • • • thioester bond with C-terminal Gly of ubiquitin Ubiquitin is then transferred to a Cys-thiol of E 2, the ubiquitin-carrier protein Ligase (E 3) selects proteins for degradation. the E 2 -S~ubiquitin complex transfers ubiquitin to these selected proteins More than one ubiquitin may be attached to a protein target 45

46 Fig. 31 -8, p. 1034

47 Fig. 31 -8 a, p. 1034

48 Fig. 31 -8 b, p. 1034

49 Fig. 31 -8 c, p. 1034

50 Fig. 31 -9, p. 1034

51 Fig. 31 -10 a, p. 1035

52