Protein folding maturation targeting Secretory pathway signal peptide













- Slides: 41

Protein folding, maturation & targeting

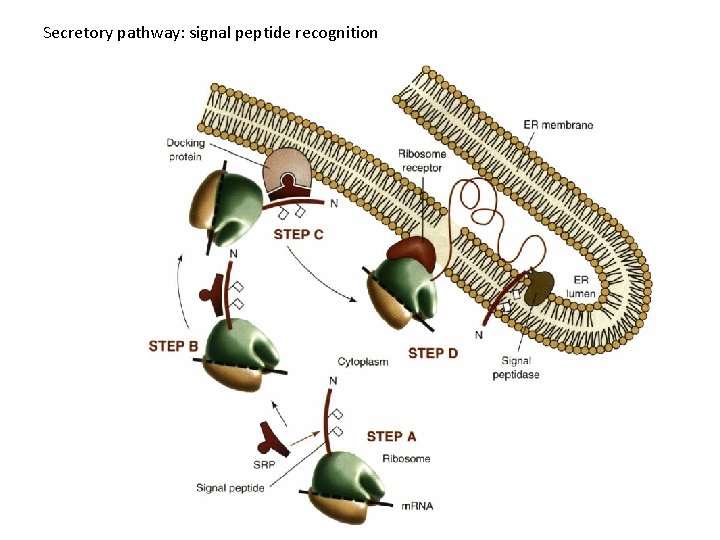
Secretory pathway: signal peptide recognition

• Glycosylation is important – alters the properties of proteins – changing their stability – solubility – act as recognition signals – influence cell-cell interactions • Glycosylation site – by the type of amino acid – its neighboring sequence in the protein – the availability of enzymes & substrates for the reactions.

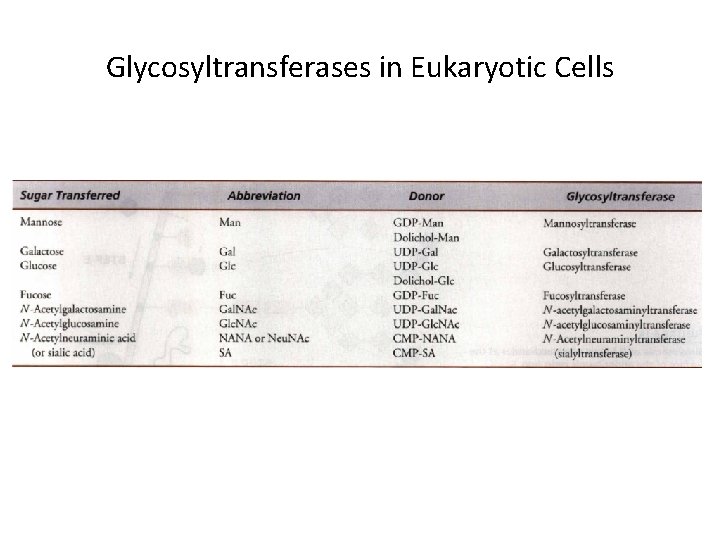
Glycosyltransferases in Eukaryotic Cells

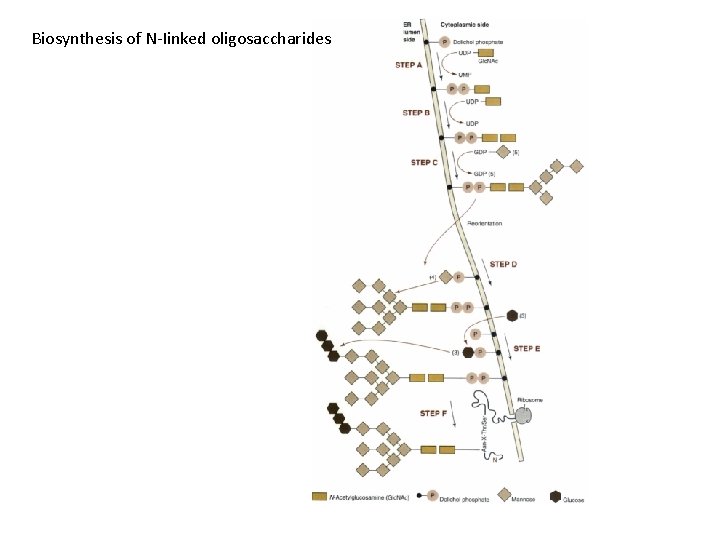
Biosynthesis of N-Iinked oligosaccharides

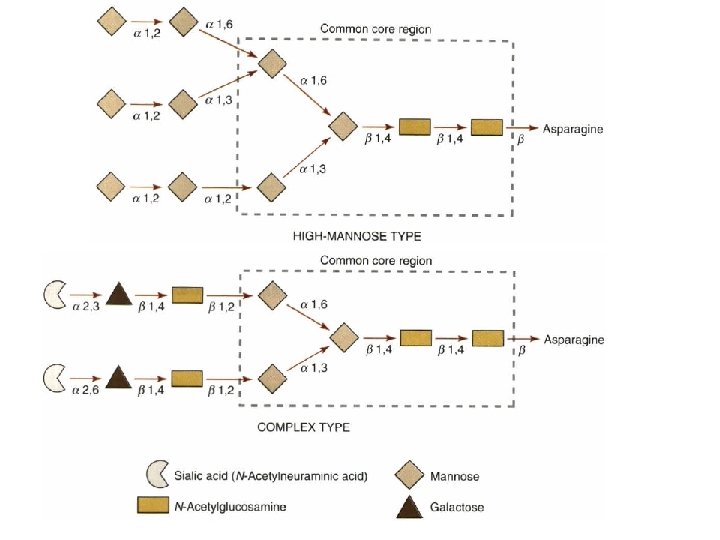
N-Iinked g. Iycosy. Iation • Asn-X-Thr/Ser • (Man)5(Glc. NAch-pyrophosphoryl-dolichol • Reorientation • Cotranslational • Glycosidases • Classes of N-linked oligosaccharides – High-mannose type – Complex type • with a larger variety of sugars and linkages • Common core region (Glc. NAc 2 Man 3)



O-linked glycosylation • O-glycosylation is posttranslational • Only residues on the protein surface serve as acceptors – (Gal. NAc-Ser/Thr) • Stepwise addition of sugars • Heterogeneity in glycoproteins is common – the types and amounts of glycosyltransferases


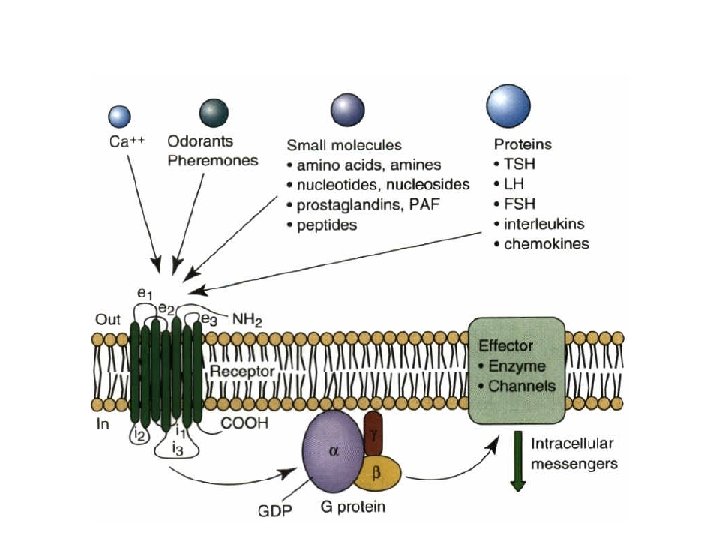
MEMBRANE AND ORGANELLE TARGETING • Protein transport uses carrier vesicles

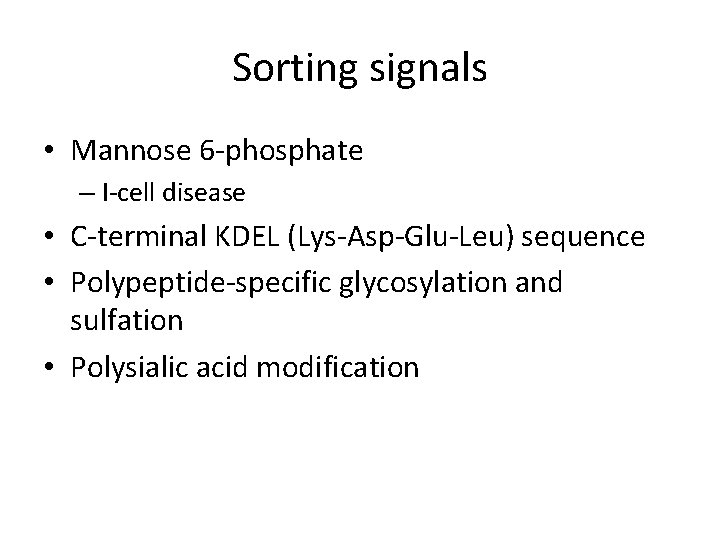
Sorting signals • Mannose 6 -phosphate – I-cell disease • C-terminal KDEL (Lys-Asp-Glu-Leu) sequence • Polypeptide-specific glycosylation and sulfation • Polysialic acid modification

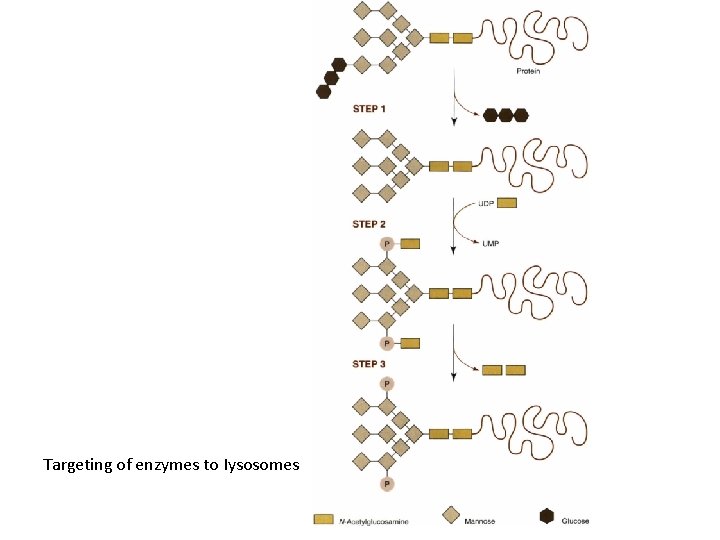
Targeting of enzymes to Iysosomes

• The secretory pathway to – Lysosomes – Plasma membrane – Secretion from the cell – Proteins of the ER and Golgi apparatus • N-terminal signal sequence • Internal signal sequence • Hydrophobic anchoring sequences

Mitochondrial proteins • N-terminal presequences • A positively charged α-helix

Nuclear Targeting • Localization signals – Clusters of basic amino acids • Peroxisome targeting – Carboxy-terminal tripeptide, Ser-Lys-Leu (SKL) – N-terminal nonapeptide • dual location – Contain two targeting signals – Gene duplication and divergence – Alternative transctiption initiation sites

Targeting • Alternative splicing • Alternative translation initiation

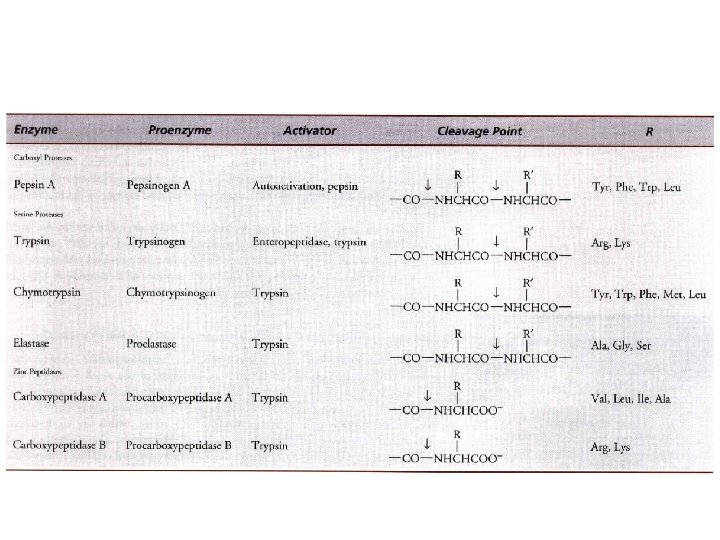
Maturation events (Posttranslational Modifications) • Some are very common – Partial proteolysis • Either end or from within – in the ER and Golgi » Insulin • others are highly restricted • Reversible modifications – regulate protein activity

• familial hyperproinsulinemia • a common means of enzyme activation – Zymogen

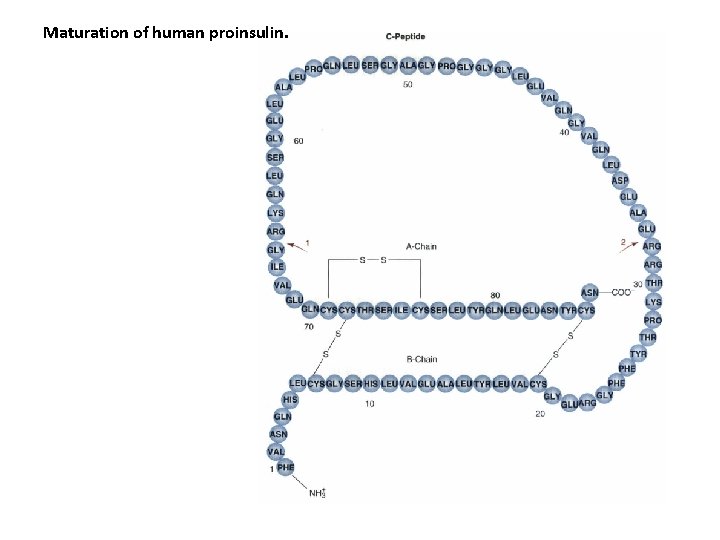
Maturation of human proinsulin.


• Amino acids can be modified after incorporation into proteins – Permanent – Reversible

• Amino-termini – Removal – Acetylarion – Alteration • Myristic or palmitic acid – G-proteins – Pyroglutamyl formation – Elongation • Disulfide bond formation – a means of localization – Cysteine modification • S-palmitoylation


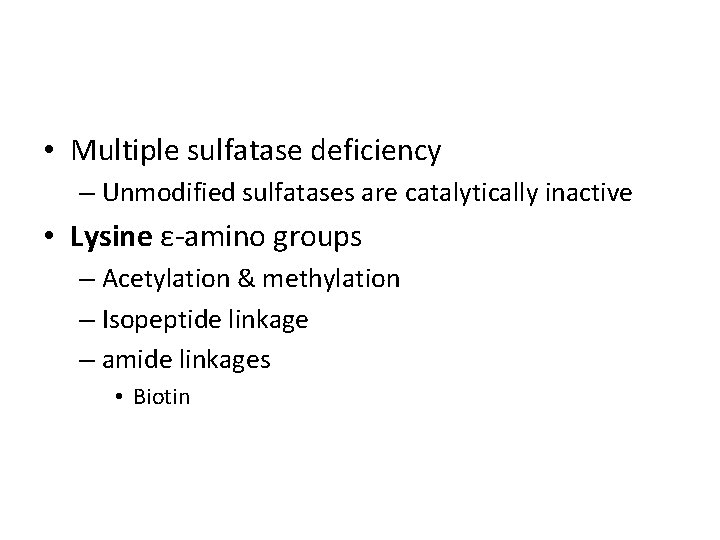
• Multiple sulfatase deficiency – Unmodified sulfatases are catalytically inactive • Lysine ε-amino groups – Acetylation & methylation – Isopeptide linkage – amide linkages • Biotin

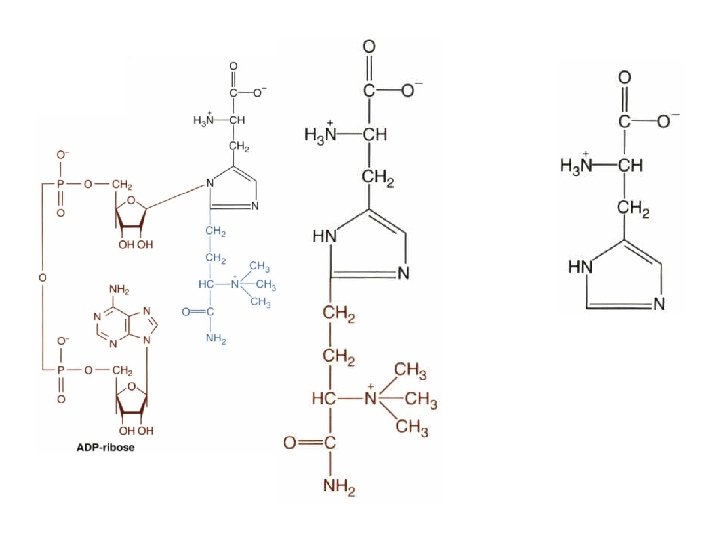
• Serine & threonine hydroxyl – Glycosylation – Phosphorylation • Tyrosine residues – Growth factor receptors – Oncogenes • Protein kinases & protein phosphatases • ADP-ribosylation on – Diphthamide – Arginine & cysteine


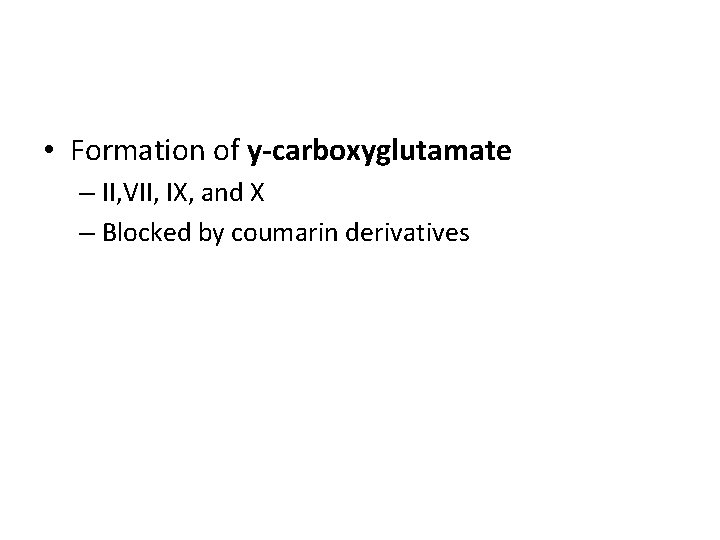
• Formation of y-carboxyglutamate – II, VII, IX, and X – Blocked by coumarin derivatives

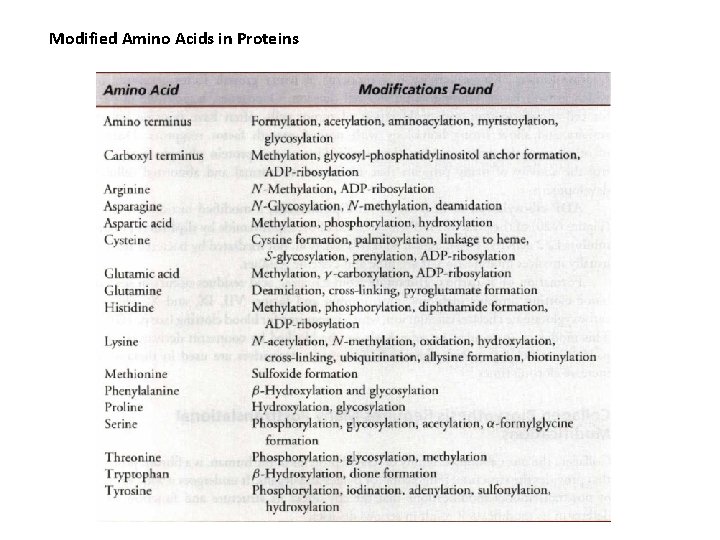
Modified Amino Acids in Proteins

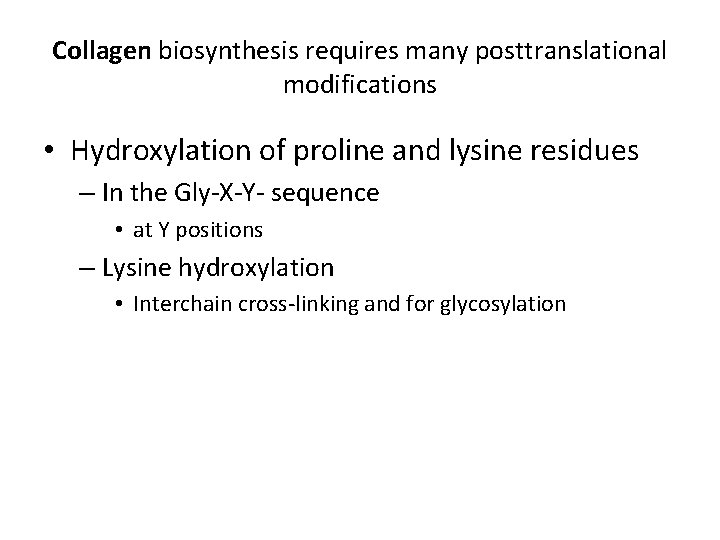
Collagen biosynthesis requires many posttranslational modifications • Hydroxylation of proline and lysine residues – In the Gly-X-Y- sequence • at Y positions – Lysine hydroxylation • Interchain cross-linking and for glycosylation

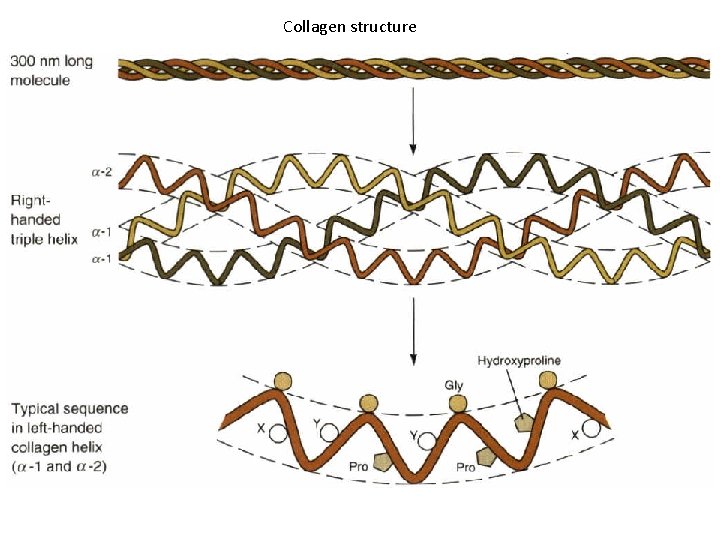
Collagen structure

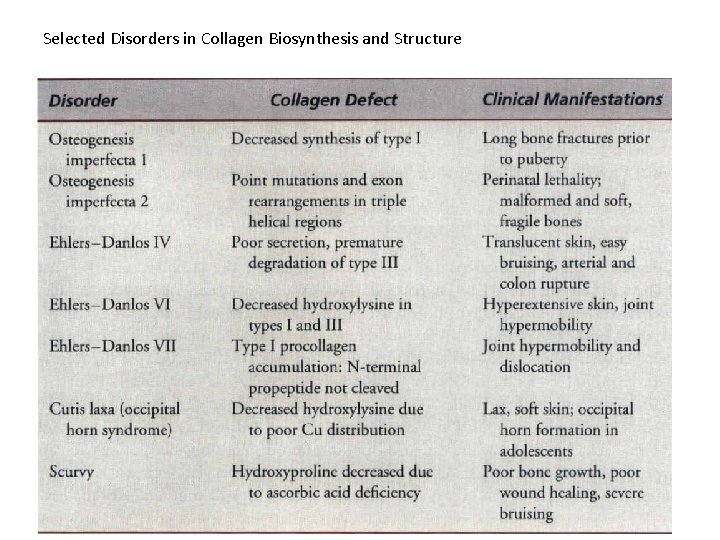
Selected Disorders in Collagen Biosynthesis and Structure

Regulation of translation • At the initiation stage – Phosphorylation of initiation factors – Global regulation • Phosphorylation of el. F-2 a. – no e. IF-2 a-GTP is available for initiation • Heme-regulated kinase • double-stranded RNA dependent kinase – Interferon • Initiation factor e. IF-4 e is activated by phosphorylation

Regulation of translation • Regulation of translation of m. RNAs – iron response element (IRE) – 5'-IRE – 3'-IRE • Polypyrimidine tract

RNA silencing and interference • Small RNA molecules – Micro-RNAs • represses translation bur does not affect m. RNA stability – Small interfering RNA (si. RNA) • Cleavage and inactivation of the target m. RNA

PROTEIN DEGRADATION AND TURNOVER

Ubiquitin-dependent proteolysis • Destabilizing PEST sequences (rich in Pro, Glu, Ser, & Thr) • Ubiquitin-interacting motif • N-end rule • Polyubiquitinylation is necessary to signal proteolysis

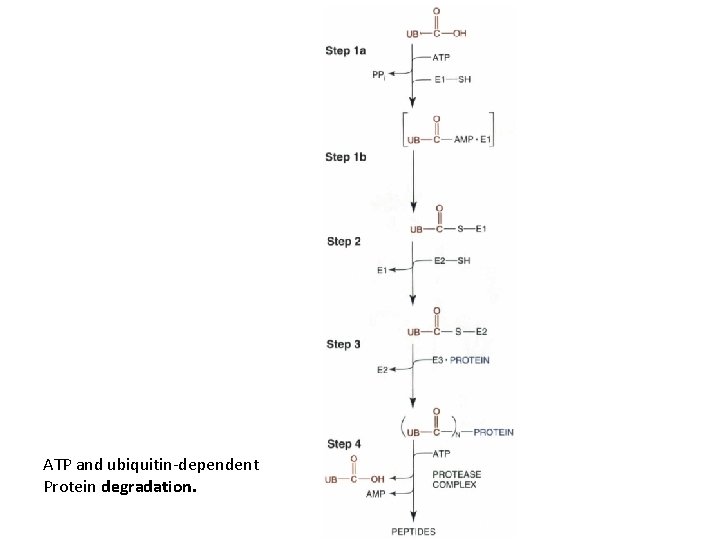
ATP and ubiquitin-dependent Protein degradation.

Model of the proteasome

• Lysosomes – from the extracellular environment – Some intracellular protein • Recognition of a specific peptide sequence

Other Proteolytic Systems • Caspases (cysteine aspartyl proteases) – Stress-induced apoptosis • thiol proteases(calpains)