Protein Design with Backbone Optimization Brian Kuhlman University

Protein Design with Backbone Optimization Brian Kuhlman University of North Carolina at Chapel Hill

Rationale for Flexible Backbone Design • Amino acid mutations often result in backbone rearrangement. • Backbone rearrangement can allow for more favorable interactions with target ligands or substrates. • Novel protein structures or complexes are generally not designable without backbone optimization.
Flexible Backbone Design Protocols in Rosetta • Design and backbone optimization of a selected region of a protein (loop or terminus) • Design and backbone optimization of a protein-protein interface • Design and backbone optimization over a whole monomeric protein
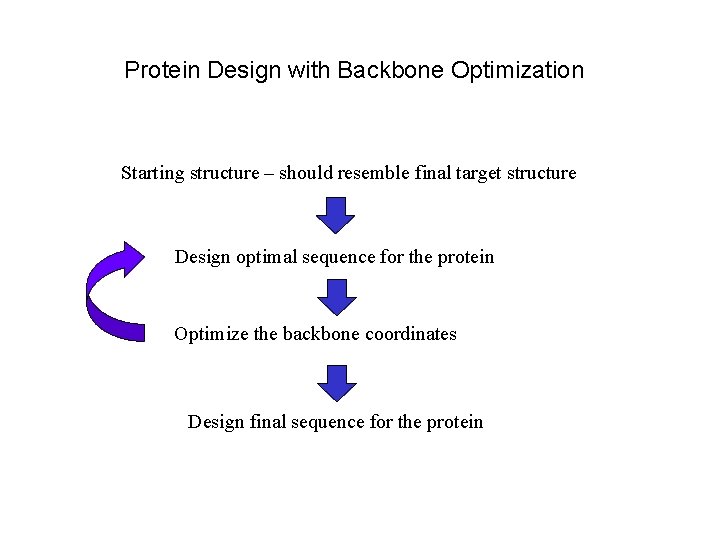
Protein Design with Backbone Optimization Starting structure – should resemble final target structure Design optimal sequence for the protein Optimize the backbone coordinates Design final sequence for the protein
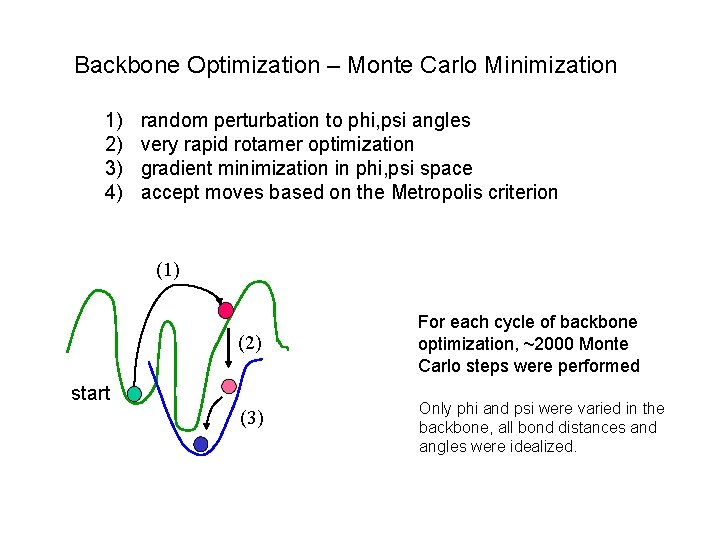
Backbone Optimization – Monte Carlo Minimization 1) 2) 3) 4) random perturbation to phi, psi angles very rapid rotamer optimization gradient minimization in phi, psi space accept moves based on the Metropolis criterion (1) (2) start (3) For each cycle of backbone optimization, ~2000 Monte Carlo steps were performed Only phi and psi were varied in the backbone, all bond distances and angles were idealized.
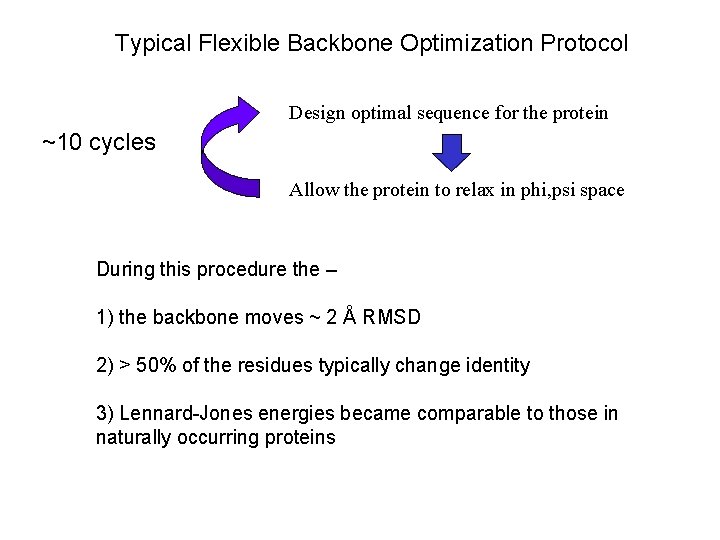
Typical Flexible Backbone Optimization Protocol Design optimal sequence for the protein ~10 cycles Allow the protein to relax in phi, psi space During this procedure the – 1) the backbone moves ~ 2 Å RMSD 2) > 50% of the residues typically change identity 3) Lennard-Jones energies became comparable to those in naturally occurring proteins

Flexible Backbone Design Protocols in Rosetta • Design and backbone optimization of a selected region of a protein (loop or terminus) • Design and backbone optimization of a protein-protein interface • Design and backbone optimization over a whole monomeric protein
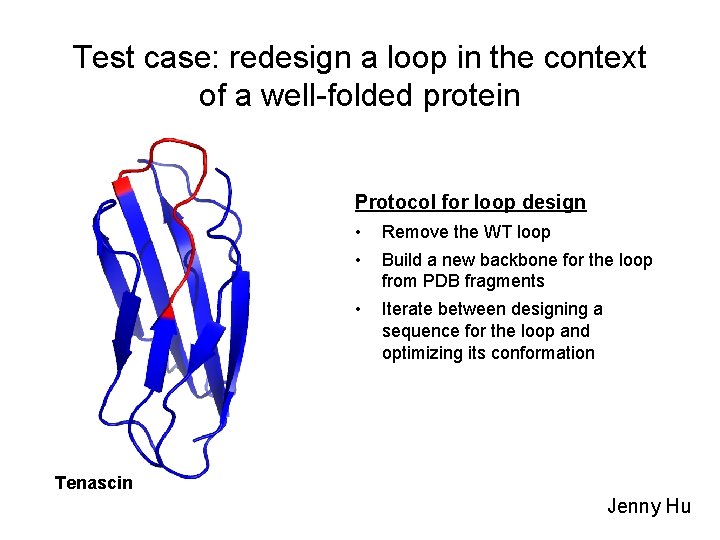
Test case: redesign a loop in the context of a well-folded protein Protocol for loop design • Remove the WT loop • Build a new backbone for the loop from PDB fragments • Iterate between designing a sequence for the loop and optimizing its conformation Tenascin Jenny Hu
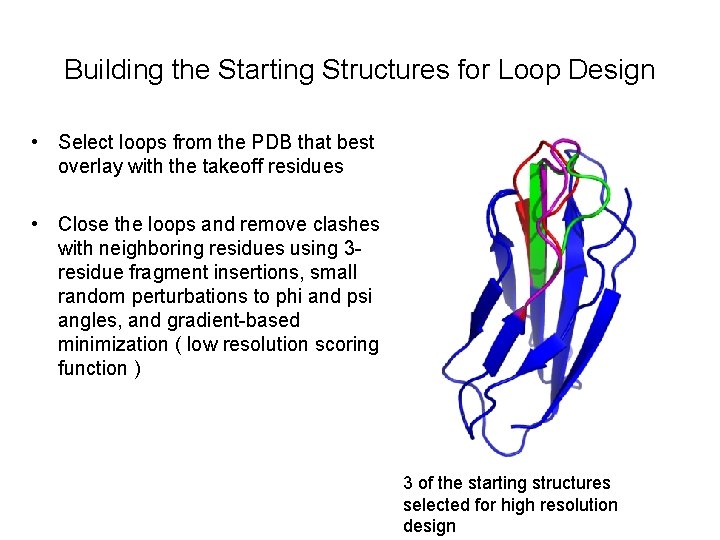
Building the Starting Structures for Loop Design • Select loops from the PDB that best overlay with the takeoff residues • Close the loops and remove clashes with neighboring residues using 3 residue fragment insertions, small random perturbations to phi and psi angles, and gradient-based minimization ( low resolution scoring function ) 3 of the starting structures selected for high resolution design
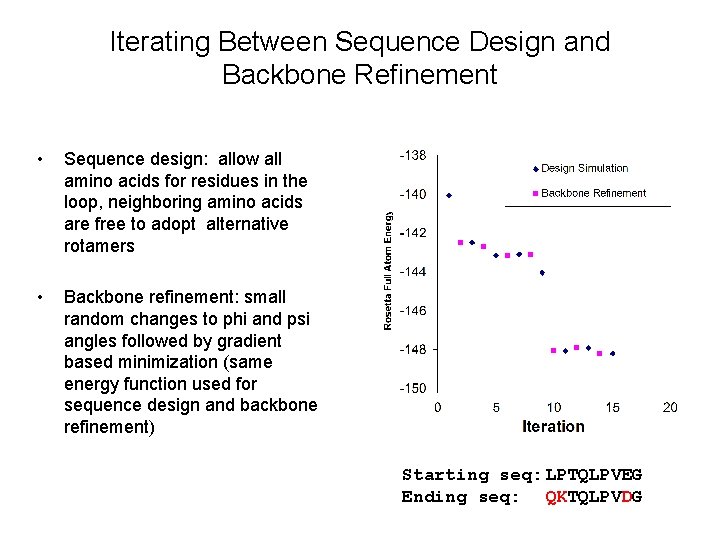
Iterating Between Sequence Design and Backbone Refinement • Sequence design: allow all amino acids for residues in the loop, neighboring amino acids are free to adopt alternative rotamers • Backbone refinement: small random changes to phi and psi angles followed by gradient based minimization (same energy function used for sequence design and backbone refinement) Starting seq: LPTQLPVEG Ending seq: QKTQLPVDG
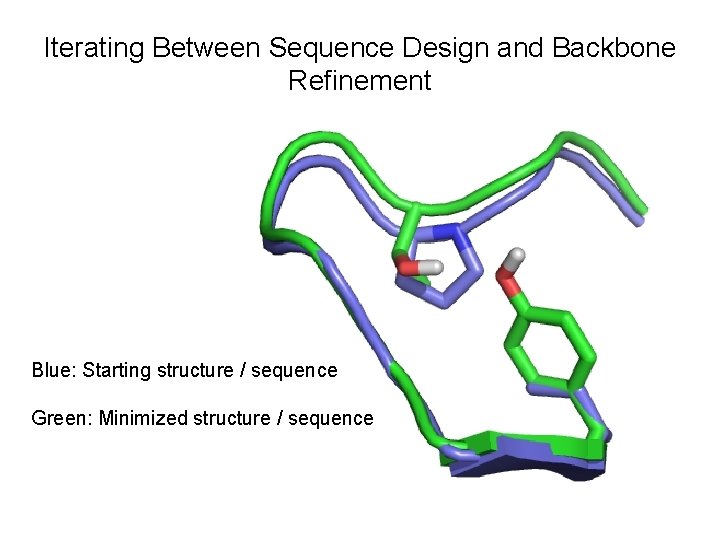
Iterating Between Sequence Design and Backbone Refinement Blue: Starting structure / sequence Green: Minimized structure / sequence
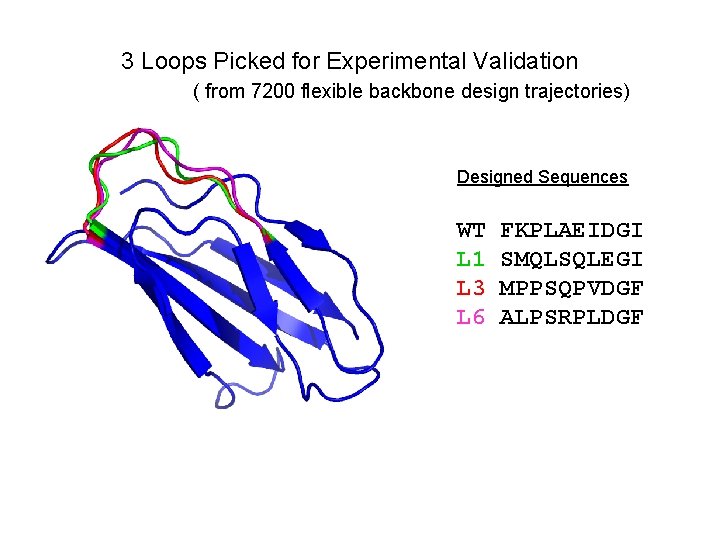
3 Loops Picked for Experimental Validation ( from 7200 flexible backbone design trajectories) Designed Sequences WT L 1 L 3 L 6 FKPLAEIDGI SMQLSQLEGI MPPSQPVDGF ALPSRPLDGF
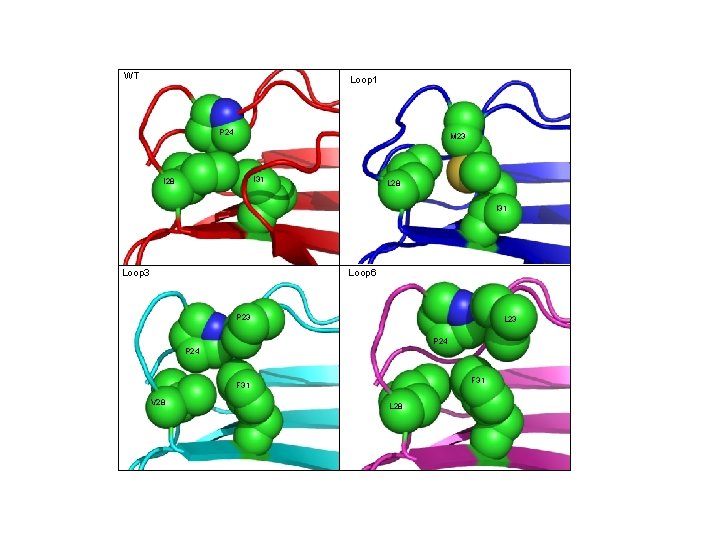
WT Loop 1 P 24 M 23 I 31 I 28 L 28 I 31 Loop 3 Loop 6 P 23 L 23 P 24 F 31 V 28 L 28
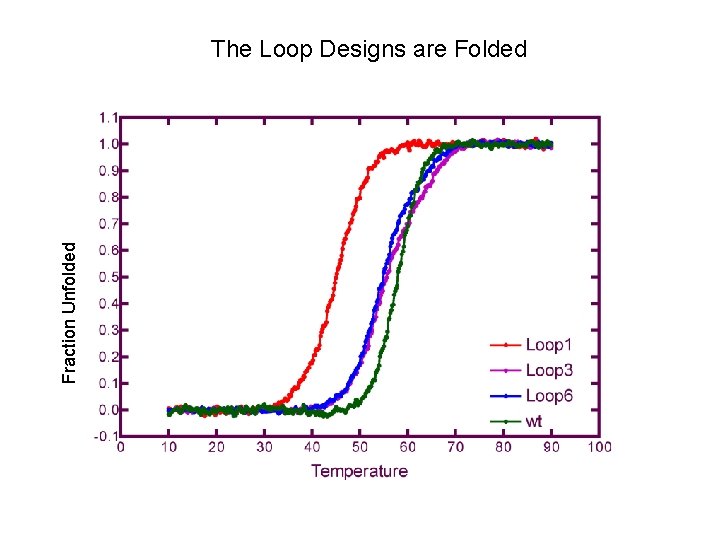
Fraction Unfolded The Loop Designs are Folded
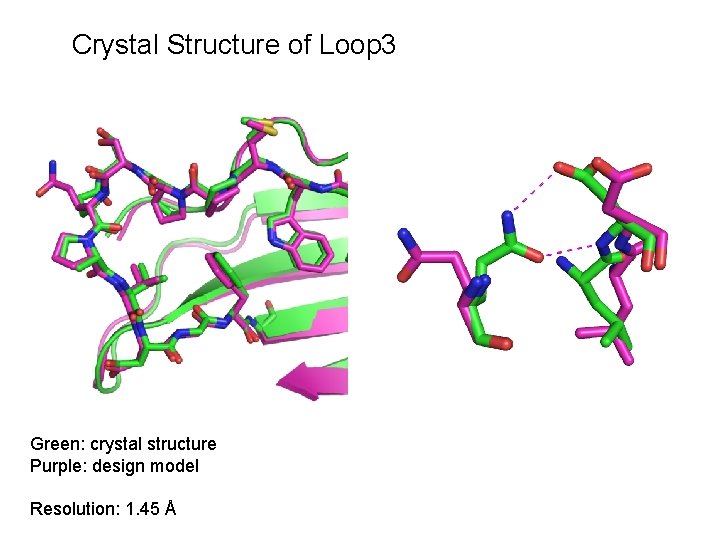
Crystal Structure of Loop 3 Green: crystal structure Purple: design model Resolution: 1. 45 Å
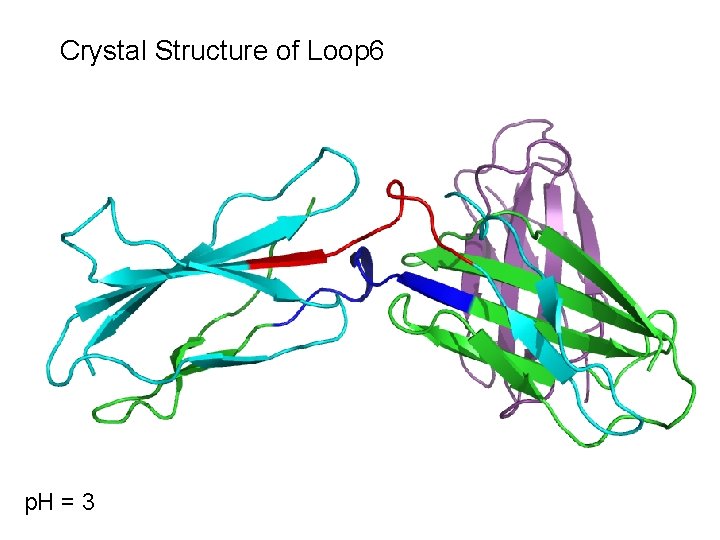
Crystal Structure of Loop 6 p. H = 3

Flexible Backbone Design Protocols in Rosetta • Design and backbone optimization of a selected region of a protein (loop or terminus) • Design and backbone optimization of a protein-protein interface • Design and backbone optimization over a whole monomeric protein
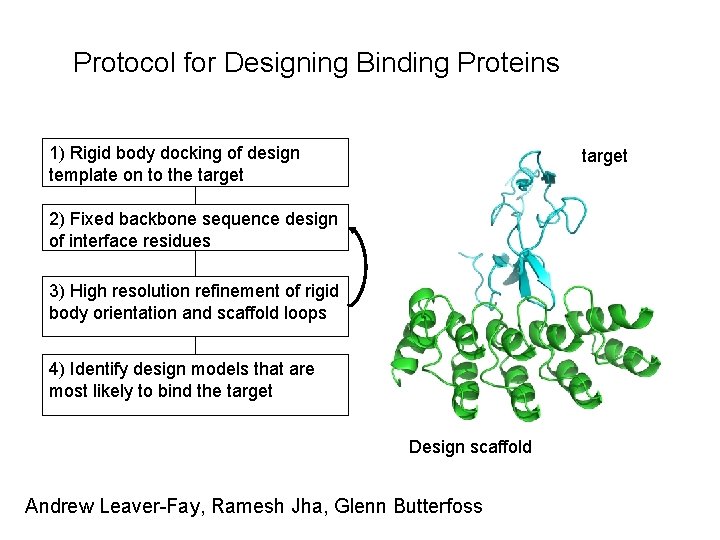
Protocol for Designing Binding Proteins 1) Rigid body docking of design template on to the target 2) Fixed backbone sequence design of interface residues 3) High resolution refinement of rigid body orientation and scaffold loops 4) Identify design models that are most likely to bind the target Design scaffold Andrew Leaver-Fay, Ramesh Jha, Glenn Butterfoss
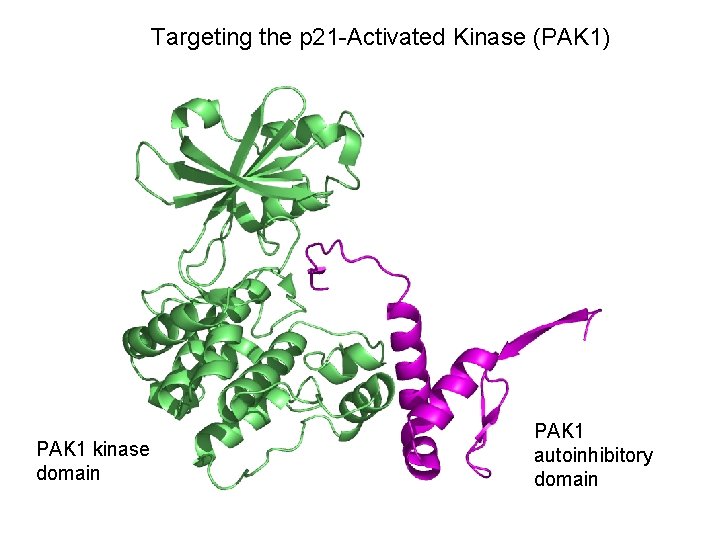
Targeting the p 21 -Activated Kinase (PAK 1) PAK 1 kinase domain PAK 1 autoinhibitory domain
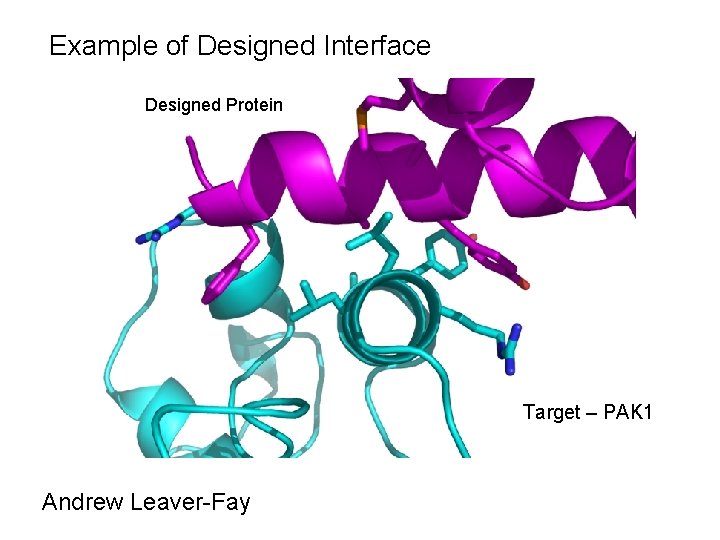
Example of Designed Interface Designed Protein Target – PAK 1 Andrew Leaver-Fay

Flexible Backbone Design Protocols in Rosetta • Design and backbone optimization of a selected region of a protein (loop or terminus) • Design and backbone optimization of a protein-protein interface • Design and backbone optimization over a whole monomeric protein
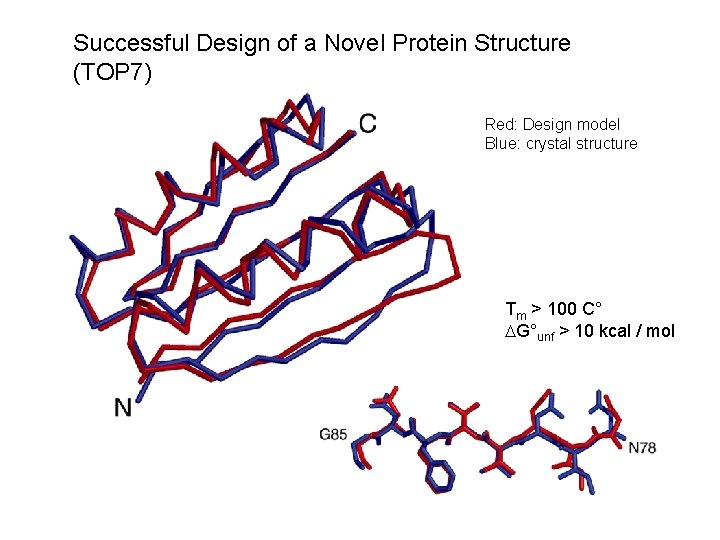
Successful Design of a Novel Protein Structure (TOP 7) Red: Design model Blue: crystal structure Tm > 100 C° DG°unf > 10 kcal / mol
Template for a b-Sandwich Protein 55 54 53 10 11 56 34 57 33 80 58 79 52 35 32 59 78 9 12 51 36 31 60 77 8 13 50 37 30 6 1 76 7 14 49 38 29 62 75 6 15 48 39 28 63 74 5 16 47 40 27 64 73 4 17 41 26 65 72 3 18 42 25 66 71 24 67 70 2 1 N 46 45 44 19 20 21 43 22 23 68 69 C
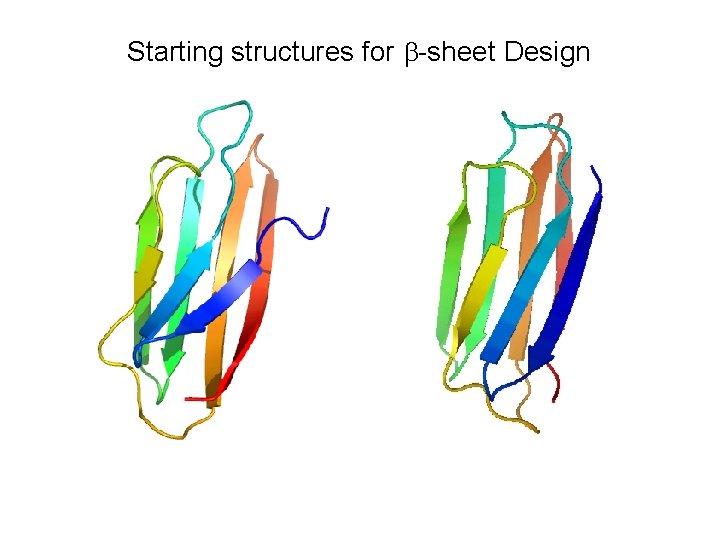
Starting structures for b-sheet Design
Current Status of b-sheet De Novo Design Project 4 sequences selected for experimental study from ~50, 000 flexible backbone simulations • All of them appear to adopt b-structure as evidenced by circular dichroism • NMR lines are broad • Gel filtration indicates that they are not monomeric

What is missing from the b-sheet design process? • Do we need to do more conformational sampling to find a backbone that is designable (positive design)? • Do we need to explicitly destabilize alternative backbone structures (negative design)?
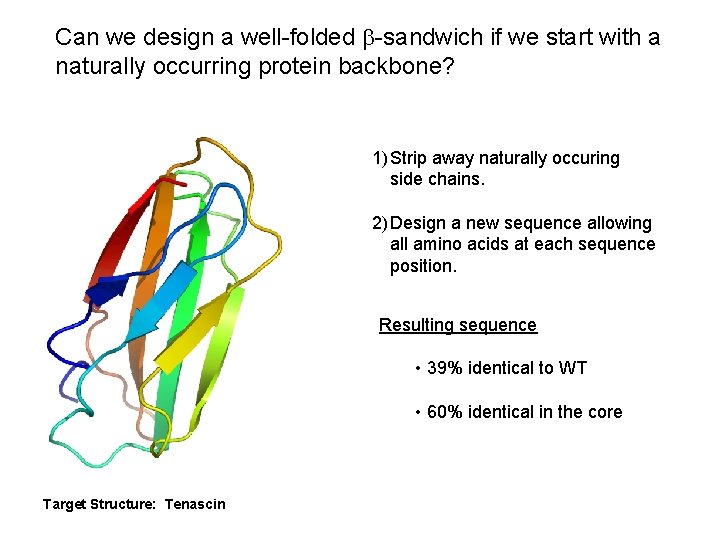
Can we design a well-folded b-sandwich if we start with a naturally occurring protein backbone? 1) Strip away naturally occuring side chains. 2) Design a new sequence allowing all amino acids at each sequence position. Resulting sequence • 39% identical to WT • 60% identical in the core Target Structure: Tenascin
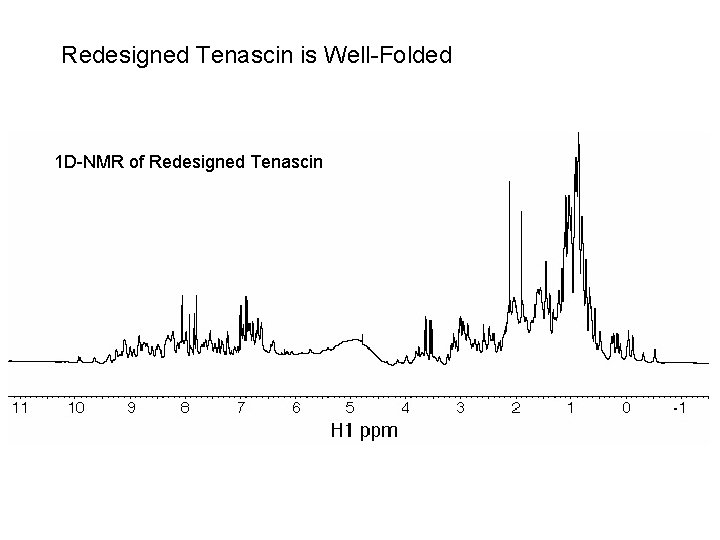
Redesigned Tenascin is Well-Folded 1 D-NMR of Redesigned Tenascin
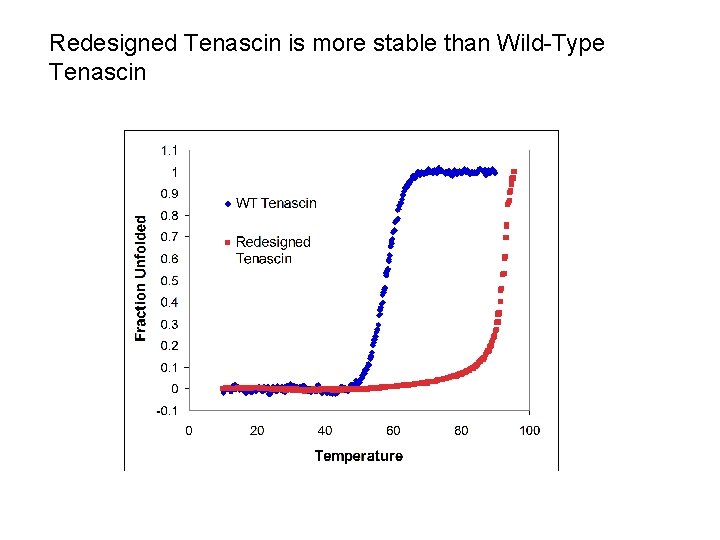
Redesigned Tenascin is more stable than Wild-Type Tenascin
Acknowledgements Loop Design Jenny Hu Hengming Ke Interface Design Andrew Leaver-Fay Glenn Butterfoss Ramesh Jha b-sheet Design Jenny Hu
- Slides: 30