Protein 3 Dstructure analysis Exercises Practicals Find update







- Slides: 22

Protein 3 D-structure analysis Exercises

Practicals Find update frequency for RCSB PDB: weekly. When was the last update? How many protein structures are there in ww. PDB? http: //www. rcsb. org/pdb/home. do http: //www. ebi. ac. uk/pdbe/#m=0&h=0&e=0&r=0&l=0&a=0&w=0

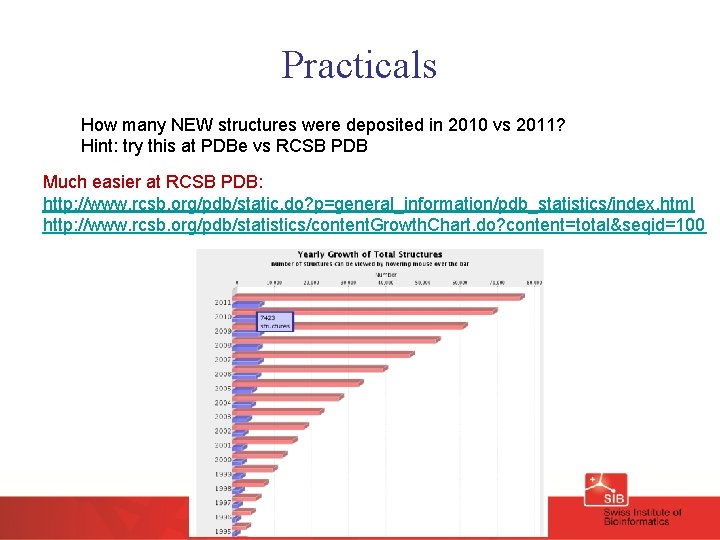
Practicals How many NEW structures were deposited in 2010 vs 2011? Hint: try this at PDBe vs RCSB PDB Much easier at RCSB PDB: http: //www. rcsb. org/pdb/static. do? p=general_information/pdb_statistics/index. html http: //www. rcsb. org/pdb/statistics/content. Growth. Chart. do? content=total&seqid=100

Practicals How many structures determined by X-ray crystallography, NMR, Electron microscopy? Use Advanced Query at RCSB PDB

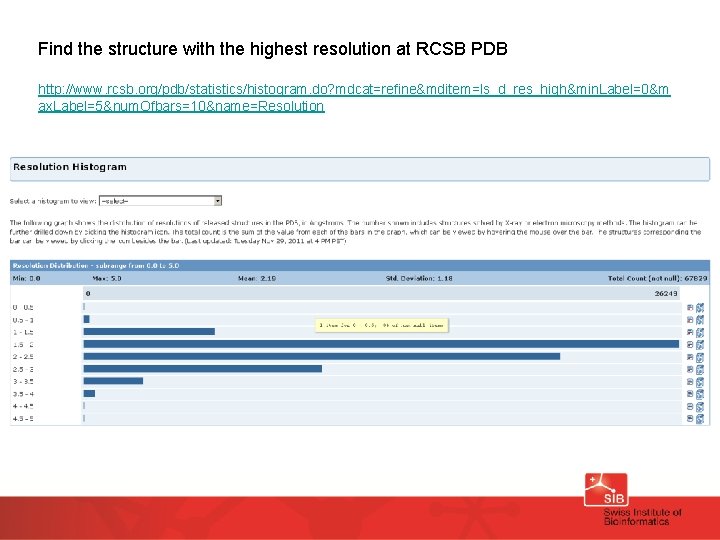
Find the structure with the highest resolution at RCSB PDB http: //www. rcsb. org/pdb/statistics/histogram. do? mdcat=refine&mditem=ls_d_res_high&min. Label=0&m ax. Label=5&num. Ofbars=10&name=Resolution

How many structures determined by X-ray crystallography, NMR, Electron microscopy? Use advanced Query at PDBe http: //www. ebi. ac. uk/pdbe-srv/view/search? search_type=advanced

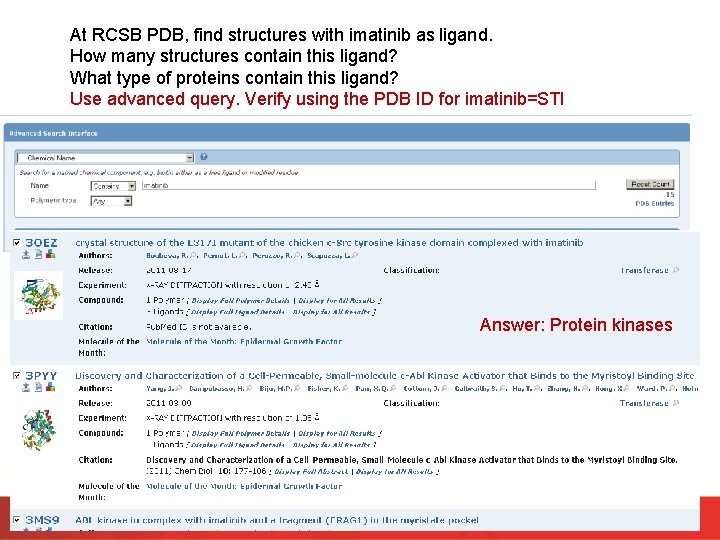
At RCSB PDB, find structures with imatinib as ligand. How many structures contain this ligand? What type of proteins contain this ligand? Use advanced query. Verify using the PDB ID for imatinib=STI Answer: Protein kinases

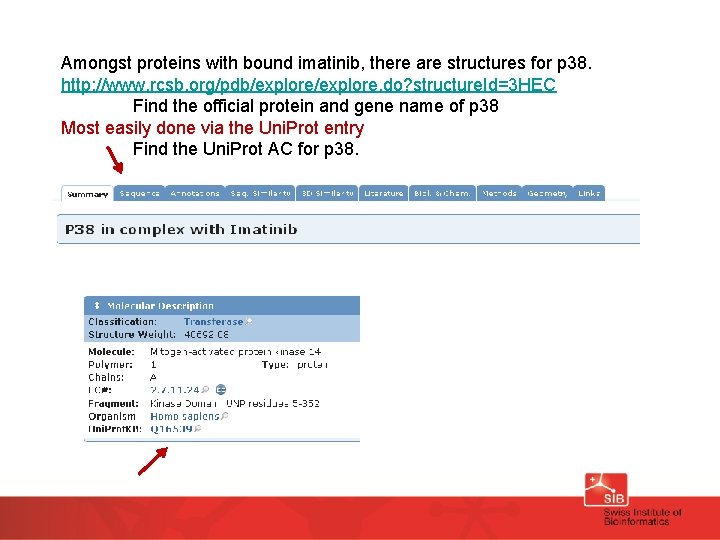
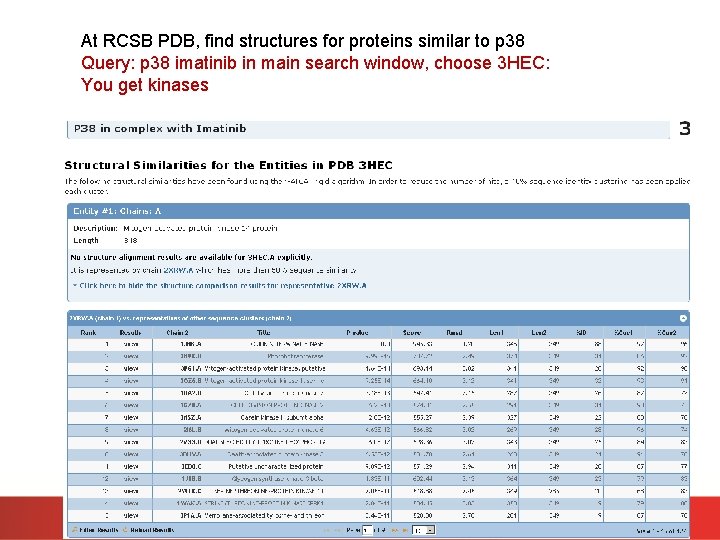
Amongst proteins with bound imatinib, there are structures for p 38. http: //www. rcsb. org/pdb/explore. do? structure. Id=3 HEC Find the official protein and gene name of p 38 Most easily done via the Uni. Prot entry Find the Uni. Prot AC for p 38.

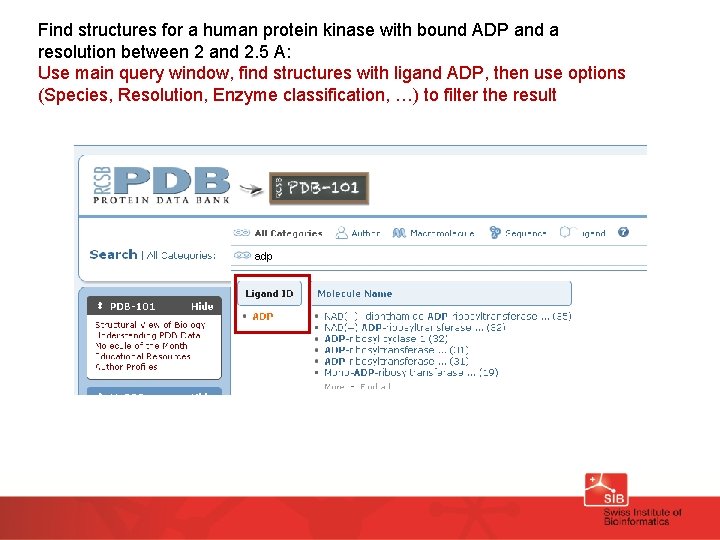
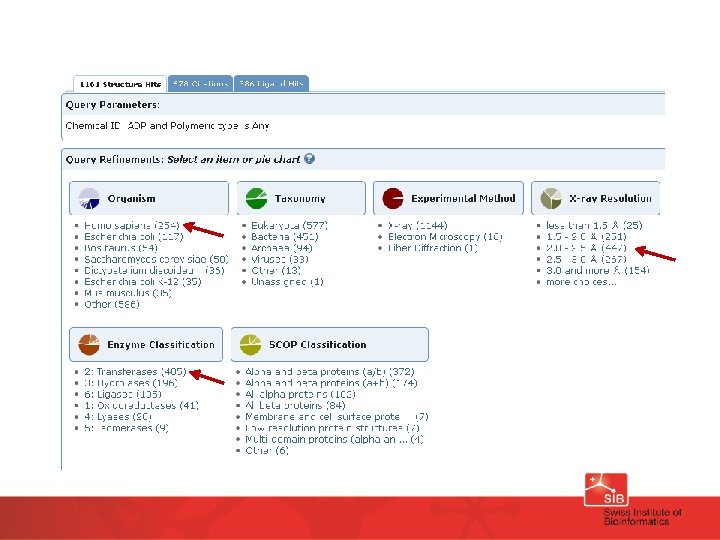
Find structures for a human protein kinase with bound ADP and a resolution between 2 and 2. 5 A: Use main query window, find structures with ligand ADP, then use options (Species, Resolution, Enzyme classification, …) to filter the result


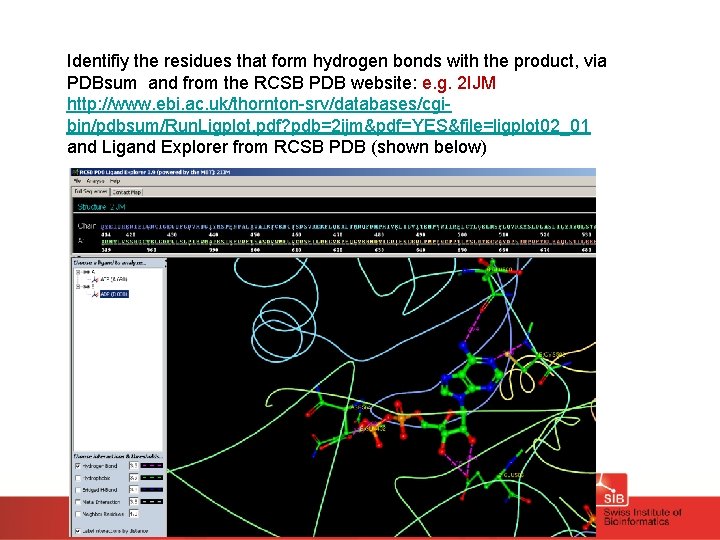
Identifiy the residues that form hydrogen bonds with the product, via PDBsum and from the RCSB PDB website: e. g. 2 IJM http: //www. ebi. ac. uk/thornton-srv/databases/cgibin/pdbsum/Run. Ligplot. pdf? pdb=2 ijm&pdf=YES&file=ligplot 02_01 and Ligand Explorer from RCSB PDB (shown below)

At RCSB PDB, find structures for proteins similar to p 38 Query: p 38 imatinib in main search window, choose 3 HEC: You get kinases

find structures for proteins similar to p 38 via the DALI database (FSSP link on PDBsum page). What kind of proteins do you recover? http: //ekhidna. biocenter. helsinki. fi/dali_server/results/3 hec/index. html

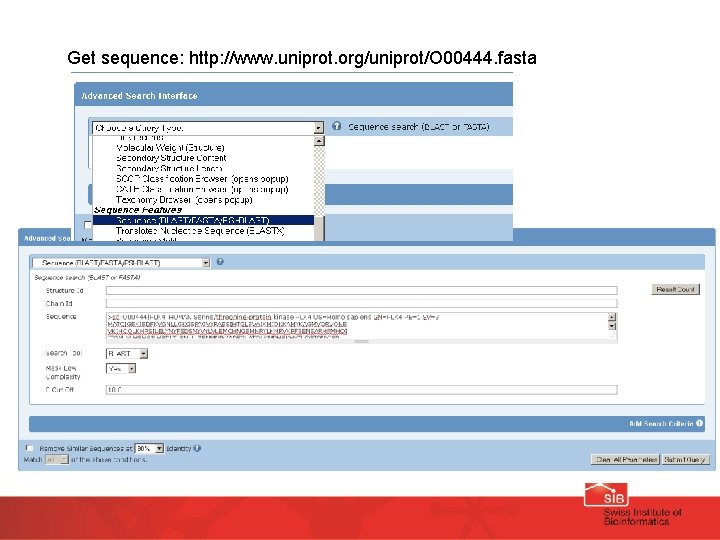
Use the sequence of human PLK 4 to find a matching structure (advanced search tools) Sequence: http: //www. uniprot. org/uniprot/O 00444. fasta

Get sequence: http: //www. uniprot. org/uniprot/O 00444. fasta

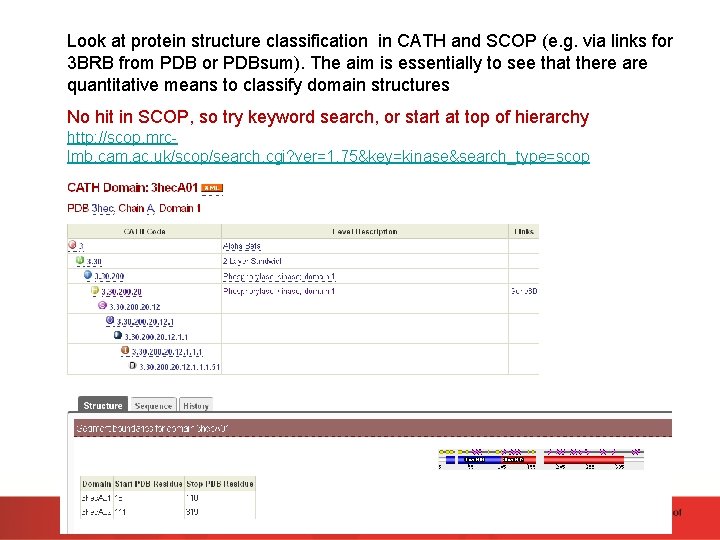
Look at protein structure classification in CATH and SCOP (e. g. via links for 3 BRB from PDB or PDBsum). The aim is essentially to see that there are quantitative means to classify domain structures No hit in SCOP, so try keyword search, or start at top of hierarchy http: //scop. mrclmb. cam. ac. uk/scop/search. cgi? ver=1. 75&key=kinase&search_type=scop

How many PDB structures are represented in CATH and in SCOP? What is the reason for the difference relative to the total number of experimental structures? Check their home pages: http: //www. cathdb. info/ http: //scop. mrc-lmb. cam. ac. uk/scop/index. html More automated procedures can handle more data – human beings can handle less data, but using your brains adds extra value

Starting with the structure for human FTO, find other proteins with similar 3 D-structure via PDB http: //www. rcsb. org/pdb/explore/structure. Cluster. do? structure. Id=3 LFM or DALI (FSSP) : http: //ekhidna. biocenter. helsinki. fi/dali_server/results/3 lfm/index. html What kind of proteins do you find (function, taxonomy)? Answer: dioxygenases, more specifically ALPHA-KETOGLUTARATEDEPENDENT DIOXYGENASE Taxons: Mammalia (alk. B homologs), and bacteria (alk. B from E. coli) What are their ligands? Answer: alpha-ketoglutarate and iron http: //www. rcsb. org/pdb/explore. do? structure. Id=2 IUW

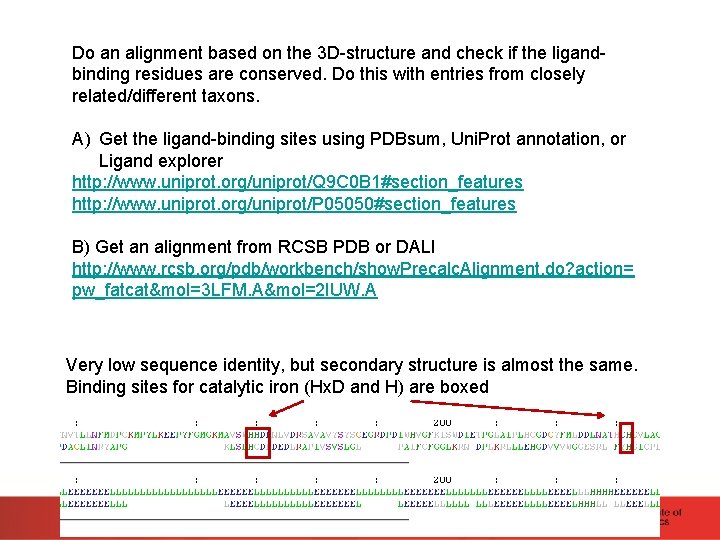
Do an alignment based on the 3 D-structure and check if the ligandbinding residues are conserved. Do this with entries from closely related/different taxons. A) Get the ligand-binding sites using PDBsum, Uni. Prot annotation, or Ligand explorer http: //www. uniprot. org/uniprot/Q 9 C 0 B 1#section_features http: //www. uniprot. org/uniprot/P 05050#section_features B) Get an alignment from RCSB PDB or DALI http: //www. rcsb. org/pdb/workbench/show. Precalc. Alignment. do? action= pw_fatcat&mol=3 LFM. A&mol=2 IUW. A Very low sequence identity, but secondary structure is almost the same. Binding sites for catalytic iron (Hx. D and H) are boxed

Can you get the same proteins, doing a normal BLAST search with FTO? Answer: No, they are too different Have a look at the domain structure (Inter. Pro) of FTO and the proteins that you identified. Compare Inter. Pro lines: they are different http: //www. uniprot. org/uniprot/P 05050#section_x-ref http: //www. uniprot. org/uniprot/Q 9 C 0 B 1#section_x-ref

Find 3 D-structures for transthyretin at RCSB PDB, then look for proteins with similar 3 D-structure http: //www. rcsb. org/pdb/explore. do? structure. Id=1 THC Similar structure: 2 H 6 U What is the function of transthyretin, respectively of other proteins with similar structure? Have a look at the corresponding Uni. Prot entries! How are they classified at CATH / at SCOP? Use text search http: //scop. mrc-lmb. cam. ac. uk/scop/data/scop. b. c. d. e. b. b. html http: //www. cathdb. info/cathnode/2. 60. 40. 180 NB: the proteins are too similar to be distinguished by these means

More things to do (facultative) Find protein structures with aspirin as ligand Find the chemical structure for aspirin Get all structures with the ligand aspirin. How many are there? - Go to CHEBI, get Smiles for aspirin - http: //www. ebi. ac. uk/chebi/advanced. Search. FT. do - http: //www. ebi. ac. uk/chebi/search. Id. do; E 5 D 46 C 3 E 2363 F 473 FCE 4 C 2 01 CC 2078 A 6? chebi. Id=CHEBI: 15365 - Go to RCSB PDB, use advanced search (smiles) - You can also try and draw “aspirin” using CHEBI