Protein 3 D structure 3 o and 4


- Slides: 16

Protein 3 -D structure: 3 o and 4 o structure and protein folding.

o 3 Structure • third level of protein organization • folding of polypeptide chain causes 2 o structures to interact • formation of motifs and domains

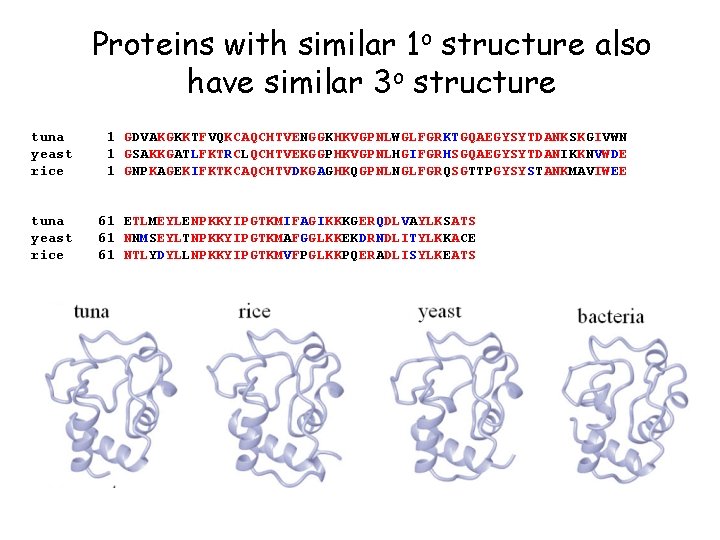
Proteins with similar 1 o structure also have similar 3 o structure tuna yeast rice 1 GDVAKGKKTFVQKCAQCHTVENGGKHKVGPNLWGLFGRKTGQAEGYSYTDANKSKGIVWN 1 GSAKKGATLFKTRCLQCHTVEKGGPHKVGPNLHGIFGRHSGQAEGYSYTDANIKKNVWDE 1 GNPKAGEKIFKTKCAQCHTVDKGAGHKQGPNLNGLFGRQSGTTPGYSYSTANKMAVIWEE 61 ETLMEYLENPKKYIPGTKMIFAGIKKKGERQDLVAYLKSATS 61 NNMSEYLTNPKKYIPGTKMAFGGLKKEKDRNDLITYLKKACE 61 NTLYDYLLNPKKYIPGTKMVFPGLKKPQERADLISYLKEATS

Common Motifs Motif • Helix-loop-helix • Coiled-coil • Helix bundle Composition all alpha-helix • Beta meander • Greek key all beta sheet • Beta-alpha-beta mixed alpha/beta

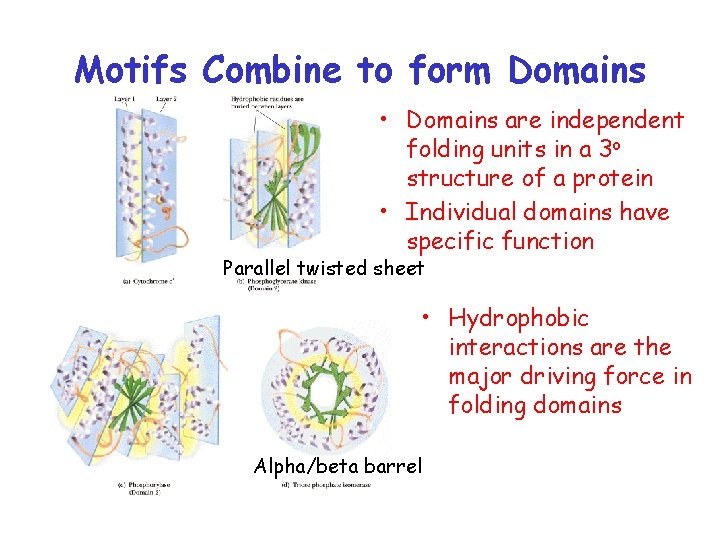
Motifs Combine to form Domains • Domains are independent folding units in a 3 o structure of a protein • Individual domains have specific function Parallel twisted sheet • Hydrophobic interactions are the major driving force in folding domains Alpha/beta barrel

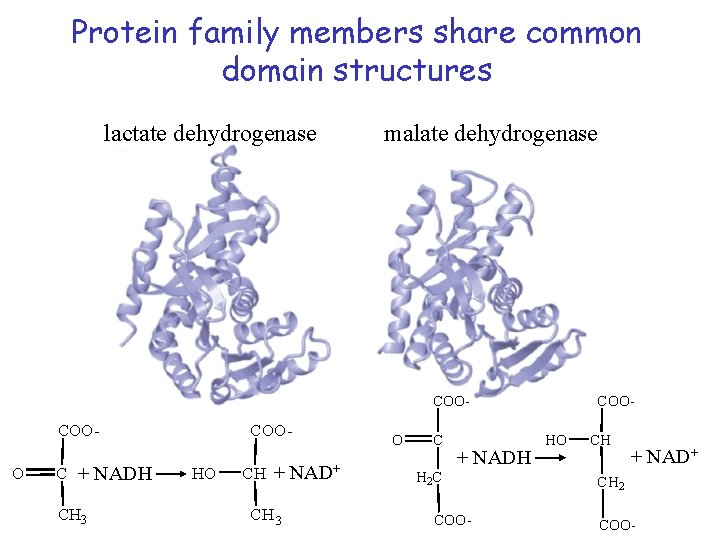
Protein family members share common domain structures lactate dehydrogenase malate dehydrogenase COO- COOO C + NADH CH 3 COOHO CH + NAD+ CH 3 O C + NADH H 2 C COO- HO CH + NAD+ CH 2 COO-

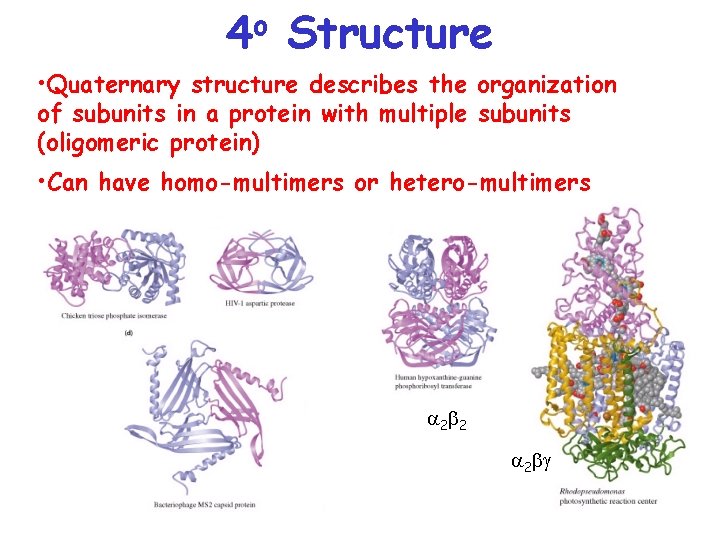
o 4 Structure • Quaternary structure describes the organization of subunits in a protein with multiple subunits (oligomeric protein) • Can have homo-multimers or hetero-multimers a 2 b 2 a 2 bg

4 o Structure • Determine molecular weight of native protein by gel permeation chromatography • Determine molecular weight of individual subunits by SDS-PAGE • Can use the information to determine subunit composition If……. Native protein – 160, 000 daltons and a-Subunit – 50, 000 daltons b-Subunit – 30, 000 daltons Then…… Protein can have a 2 b 2 structure

4 o Structure • Subunits held together by non-covalent interactions • Oligomeric protein is more stable than disassociated subunits • Active site often made up of AA residues from different subunits • 4 o and 3 o structure is often affected by ligand (substrate or inhibitor) binding. Important in enzyme regulation

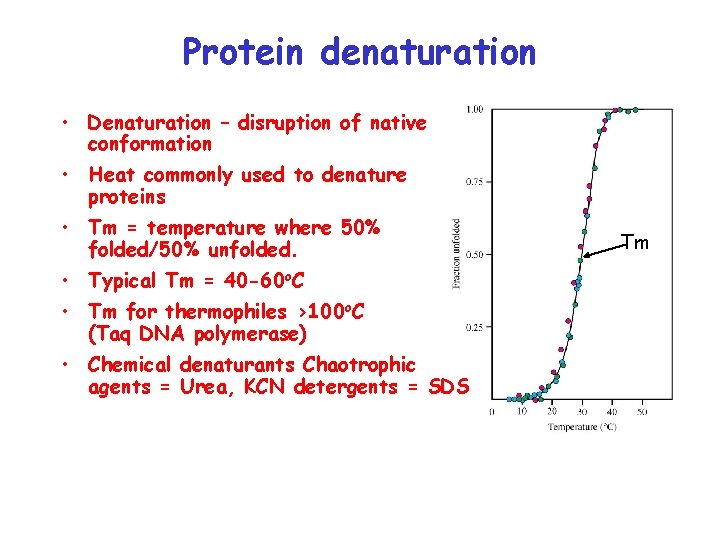
Protein denaturation • Denaturation – disruption of native conformation • Heat commonly used to denature proteins • Tm = temperature where 50% folded/50% unfolded. • Typical Tm = 40 -60 o. C • Tm for thermophiles >100 o. C (Taq DNA polymerase) • Chemical denaturants Chaotrophic agents = Urea, KCN detergents = SDS Tm

Protein Folding • Ribonuclease A (RNase A) will refold to native structure spontaneously (1 minute) • >1050 possible conformations • If 10 -13 sec per conformation would take 1030 years to sample enough to determine structure • How do proteins fold so quickly?

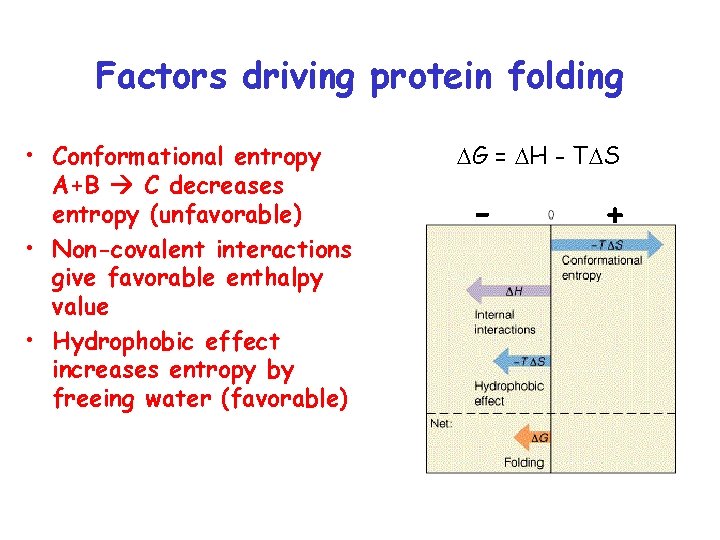
Factors driving protein folding • Conformational entropy A+B C decreases entropy (unfavorable) • Non-covalent interactions give favorable enthalpy value • Hydrophobic effect increases entropy by freeing water (favorable) DG = DH - TDS - +

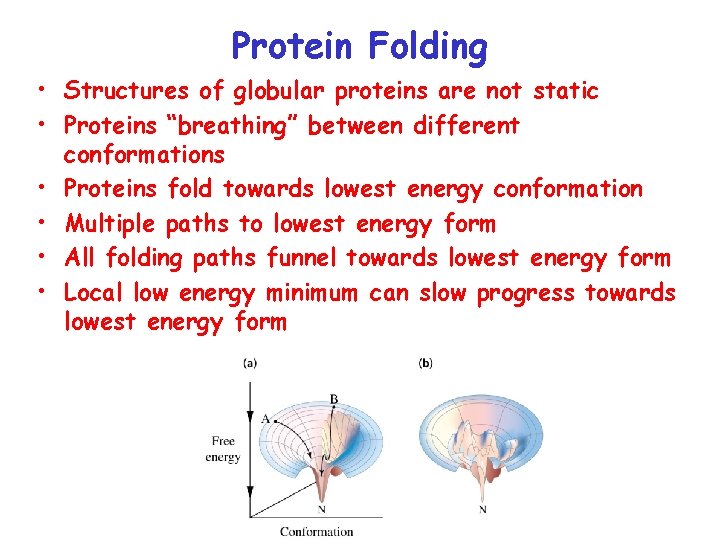
Protein Folding • Structures of globular proteins are not static • Proteins “breathing” between different conformations • Proteins fold towards lowest energy conformation • Multiple paths to lowest energy form • All folding paths funnel towards lowest energy form • Local low energy minimum can slow progress towards lowest energy form

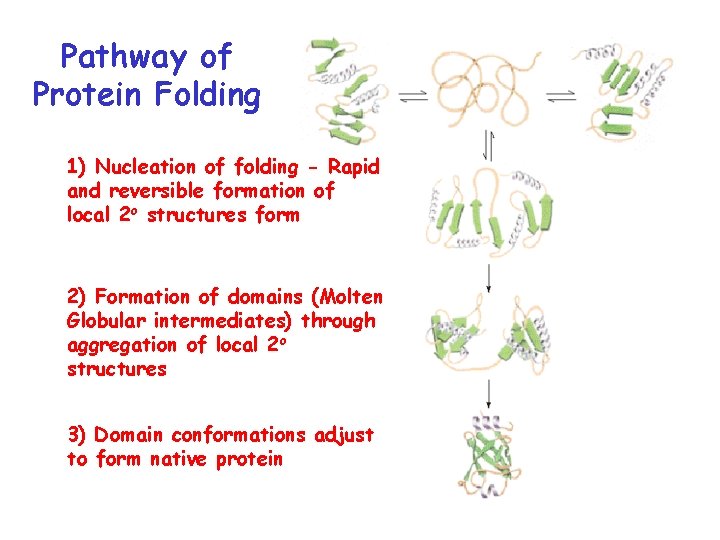
Pathway of Protein Folding 1) Nucleation of folding - Rapid and reversible formation of local 2 o structures form 2) Formation of domains (Molten Globular intermediates) through aggregation of local 2 o structures 3) Domain conformations adjust to form native protein

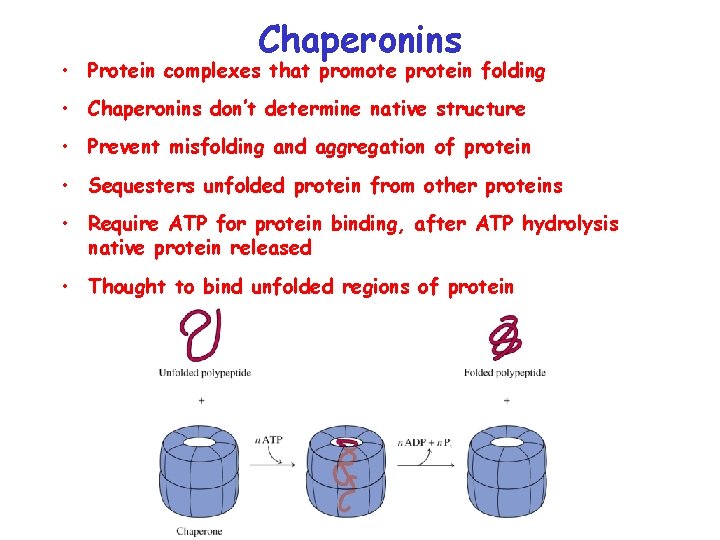
Chaperonins • Protein complexes that promote protein folding • Chaperonins don’t determine native structure • Prevent misfolding and aggregation of protein • Sequesters unfolded protein from other proteins • Require ATP for protein binding, after ATP hydrolysis native protein released • Thought to bind unfolded regions of protein

Disulfides Bonds • Stabilize native structure • Formed after native conformation achieved • Abundant in secreted proteins but not in intracellular proteins • Protein disulfide isomerase catalyzes reduction of incorrect disulfide linkages