Preparing for the Exam Question 1 Amino acid








- Slides: 28

Preparing for the Exam Question 1: Amino acid structures and abbreviations. Question 2: Mechanism of trypsin. Question 3: Serine protease evolution and function. Question 4: Non-covalent inhibition of Protein Function Question 5: Purification and analysis of a protein Question 6: Michaelis-Menton kinetics Question 7: Reaction coordinates and enzyme catalyzed reactions Question 8: Regulation of Protein Function Question 9: Protein folding Question 10: IMFs Question 11: Lysozyme mechanism Question 12: Hemoglobin and the Bohr Effect Question 13: RNAse A Mechanism

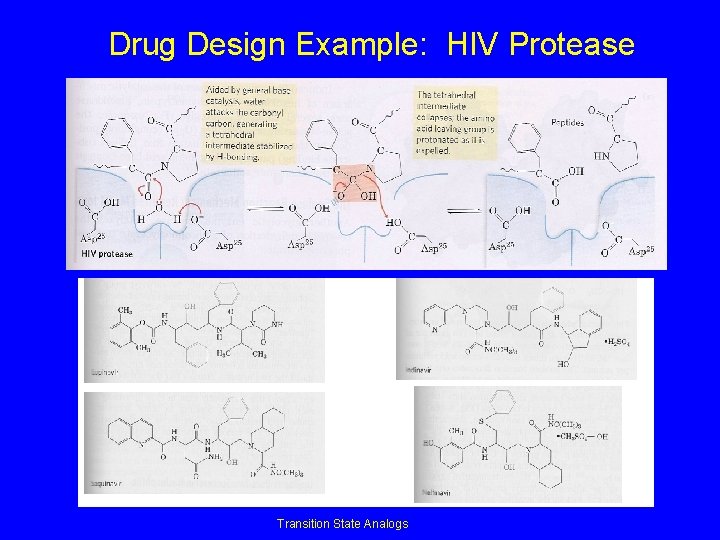
Drug Design Example: HIV Protease Transition State Analogs

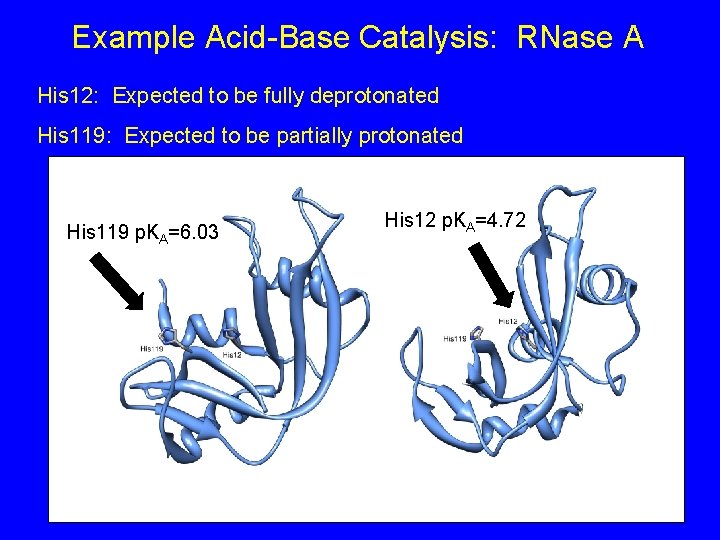
Example Acid-Base Catalysis: RNase A Enzyme responsible for degrading RNA in the cell

Example Acid. Base Catalysis: RNase A

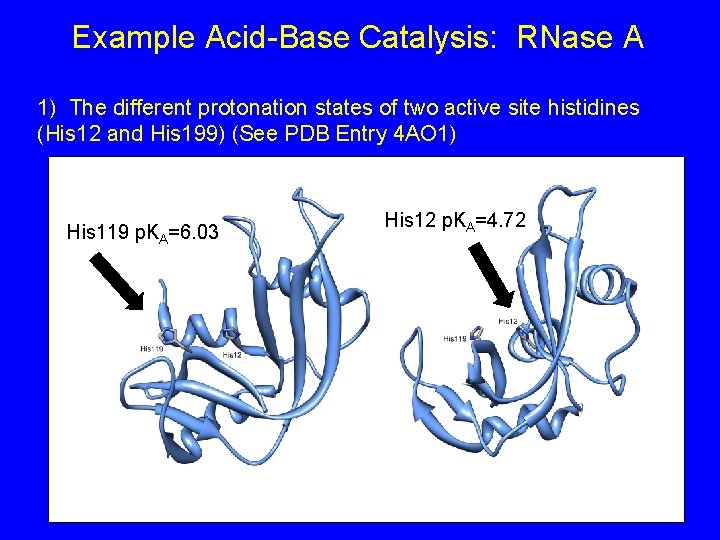
Example Acid-Base Catalysis: RNase A 1) The different protonation states of two active site histidines (His 12 and His 199) (See PDB Entry 4 AO 1) His 119 p. KA=6. 03 His 12 p. KA=4. 72

Example Acid-Base Catalysis: RNase A His 12: Expected to be fully deprotonated His 119: Expected to be partially protonated His 119 p. KA=6. 03 His 12 p. KA=4. 72

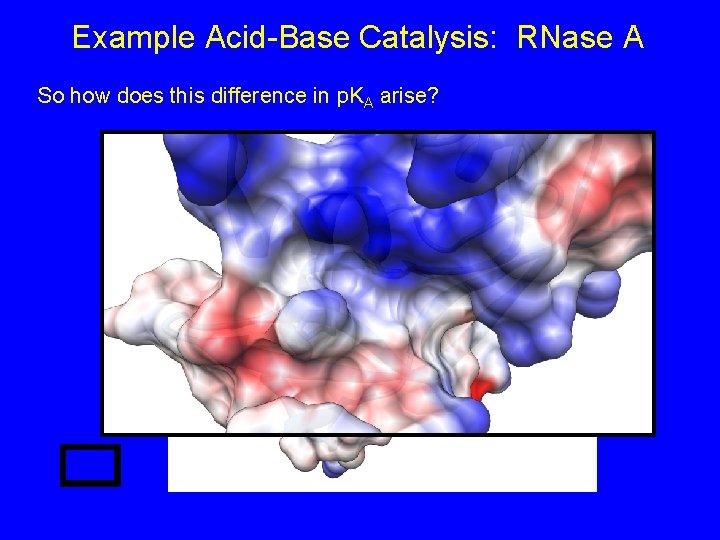
Example Acid-Base Catalysis: RNase A So how does this difference in p. KA arise?

Example Acid-Base Catalysis: RNase A 2) Substrate Binding: Orientation Effects • Look at the Coulombic surface • What is the charge of RNA? • Where would the phosphate backbone bind most productively? • Where does that binding mode place the C 2’ hydroxyl group of the 5’ ribose ring? • Which direction does the RNA substrate run in the binding site?

Example Acid-Base Catalysis: RNase A • Look at the Coulombic surface • What is the charge of RNA? • Where would the phosphate backbone bind most productively? • Where does that binding mode place the C 2’ hydroxyl group of the 5’ ribose ring? • Which direction does the RNA substrate run in the binding site? By looking at an appropriate representation of the protein and knowing the enzyme-catalyzed reaction, you are able to answer critical mechanistic questions about an enzyme you only recently knew existed

Protein Regulation There are three basic mechanisms to control/regulate protein function 1. Localization of the protein and/or its partners 2. Binding of effector molecules or Posttranslational Modifications 3. Physical amounts of protein and the lifetime of each molecule • This is the simplest mechanism to control protein function

1. Regulation by Localization Signal sequences target protein to specific compartment Covalent modifications force protein to stay in a specific compartment Interactions with other proteins that exist in a specific compartment

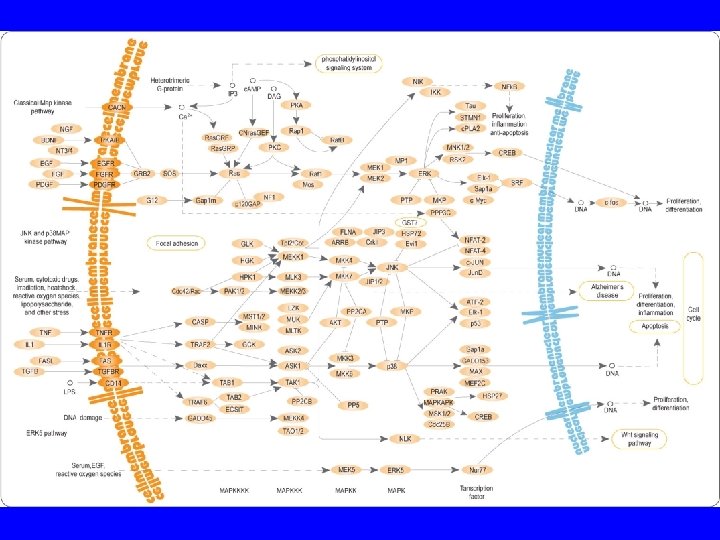
2. Regulation by Effector Molecules Usually induce conformational changes that alter protein function • Inhibitors, cooperativity Allostery • Alternate binding sites that promote or weaken subsequent binding events when occupied Covalent modifications • Phosphorylation, methylation, glycosylation, acetylation, etc.

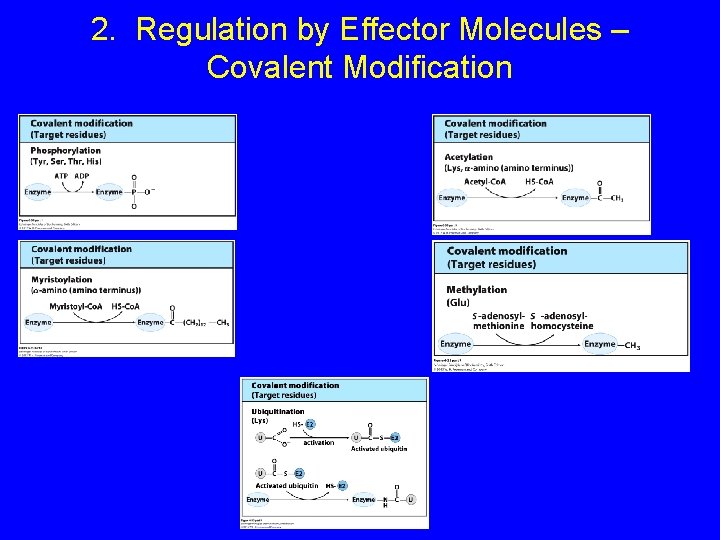
2. Regulation by Effector Molecules – Covalent Modification

3. Protein Regulation by Degradation (Lifetime) The lifetime of a protein is a simple method of regulation • Proteins involved in cell cycle or metabolism generally have short lifetimes. Why? • Structural proteins (actin/histones) have longer lifetimes. • How is a protein’s lifespan determined? • Intrinsic stability of the tertiary structure • Certain recognition sequences • Proteins are degraded in the Proteasome

1. Regulation by Localization In eukaryotic cells, proteins can be targeted to specific locales: ER, Golgi, Nucleus, mitochondrion or secreted Specific signal sequences interact with other proteins at the target site KDEL: Endoplasmic reticulum KRKR: Nucleus Hydrophobic residues: Secretion (Golgi) (Why? ) Signal sequences are not regulated, they are a property of the primary sequence

Role of Environment on Protein Function In other words: Why is Regulation by Localization important? Here’s one example: p. H changes drastically affect the ionization states of amino acid side chains May affect ligand binding if interactions depend upon electrostatic interactions May also cause dramatic effects in protein structure • Cathepsin D (Example #1) • Hemagglutinin (Example #2) • Diptheria toxin (Example #3)

Role of Environment on Protein Function #1 Cytosol Lysosomal protease p. H of lysosome? The amino terminal domain swivels due to ionization changes Exposes active site, allows binding and charges catalytic residues Lysosome

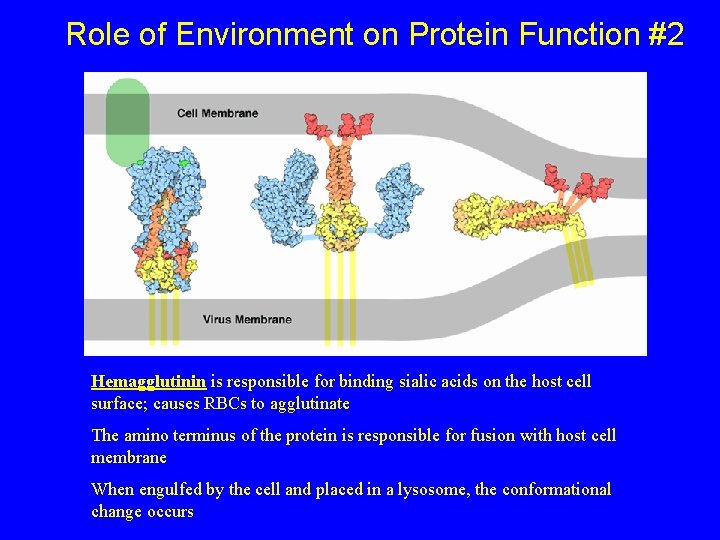
Role of Environment on Protein Function #2 Hemagglutinin is responsible for binding sialic acids on the host cell surface; causes RBCs to agglutinate The amino terminus of the protein is responsible for fusion with host cell membrane When engulfed by the cell and placed in a lysosome, the conformational change occurs

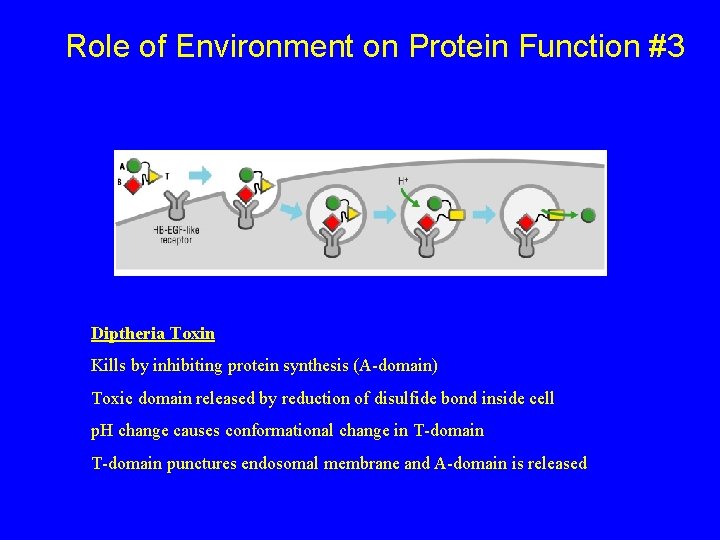
Role of Environment on Protein Function #3 Diptheria Toxin Kills by inhibiting protein synthesis (A-domain) Toxic domain released by reduction of disulfide bond inside cell p. H change causes conformational change in T-domain punctures endosomal membrane and A-domain is released

2. Regulation by Ligand Binding Simplest method to regulate a cell is to jam something into the active site In the multienzyme pathways of a cell, the product of one enzyme in a pathway may inhibit another Feedback and Feed-forward Inhibition Lots of examples available: but glycolysis is perhaps one of the best (PFK)

Cooperativity Another way to fine tune enzymatic action is to jam something into another site (not the active site) In oligomeric enzymes, the binding of a ligand may induce a conformational change that affects binding of ligand in other subunits May be positive or negative +=Hemoglobin -=G 3 P dehydrogenase Effector binds active site

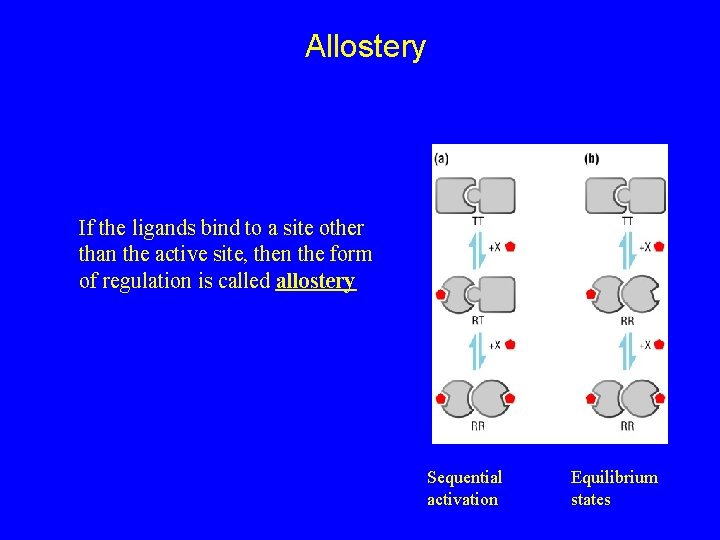
Allostery If the ligands bind to a site other than the active site, then the form of regulation is called allostery Sequential activation Equilibrium states

Post-Translational Modifications: Phosphorylation • Most common form of post-translational modification • Is Reversible • Ser, Thr and Tyr are phosphorylation targets • The large negative charge cluster of the phosphate group causes a large conformational change Reversibility granted by having 2 specific enzyme families Kinases: Phoshorylate proteins Phosphatases: Dephosphorylate proteins (hydrolysis) • 3 families that recognize each of the phosphorylated residues

Post-Translational Modifications: Phosphorylation Effects: 1) Phosphoryl oxygens hydrogen bond to mainchain amide hydrogens OR 2) Phosphoryl oxygen forms a salt-bridge with an Arginine 3) Causes conformational change as residues escape the charges or are pulled/pushed as an effect of 1) or 2) 4) Exposes areas for other proteins to interact with

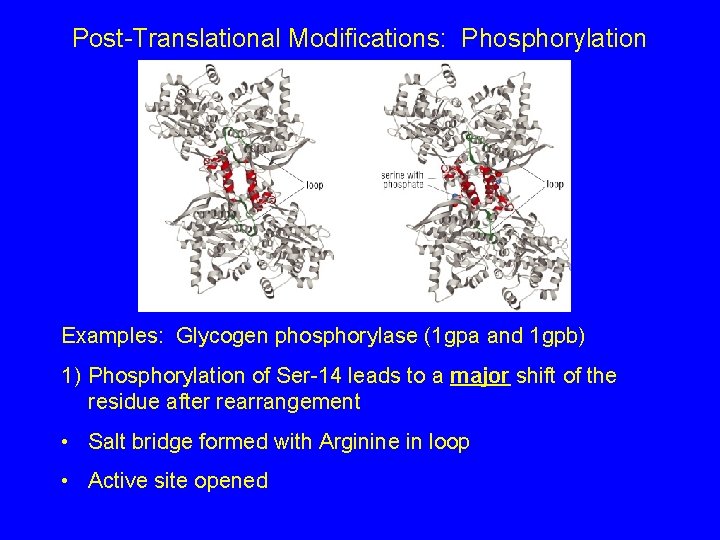
Post-Translational Modifications: Phosphorylation Examples: Glycogen phosphorylase (1 gpa and 1 gpb) 1) Phosphorylation of Ser-14 leads to a major shift of the residue after rearrangement • Salt bridge formed with Arginine in loop • Active site opened

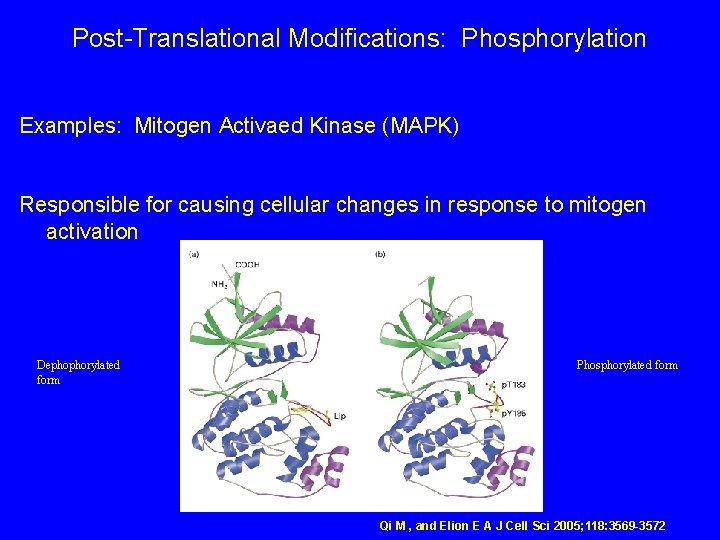
Post-Translational Modifications: Phosphorylation Examples: Mitogen Activaed Kinase (MAPK) Responsible for causing cellular changes in response to mitogen activation Dephophorylated form Phosphorylated form Qi M , and Elion E A J Cell Sci 2005; 118: 3569 -3572


Understanding the Implications of PTMs Key in signalling and marking proteins for termination Mutations may block PTM sites or create them In cancer cells, changing roles of acetylation, methylation, phosphorylation and glycosylation have major roles In parasitic infections and bites/stings/jabs from poisonous creatures, cleaving glycosyl residues and GPI anchors helps drive the infection or accelerate death/paralysis In the immune system, mutations in glycosylation pathways and/or glycosylation targets have dire consequences