PREMIER Biosoft Accelerating Research in Life Sciences Glycan
PREMIER Biosoft Accelerating Research in Life Sciences

Glycan quantitation using Thermo Scientific™ aminoxy. TMT™ (Tandem Mass Tag™) Label Reagents

Topics to be explained New Project creation Loading Thermo Scientific native. raw file format High Throughput glycan identification using aminoxy. TMT 6 as reducing end modification Model experiment design assigning the reporter ion channels to different biological samples Reporting of absolute/ normalized glycan abundance Multiple charts and plots depicting the glycan expressions across different Biological samples Portable Report Generation
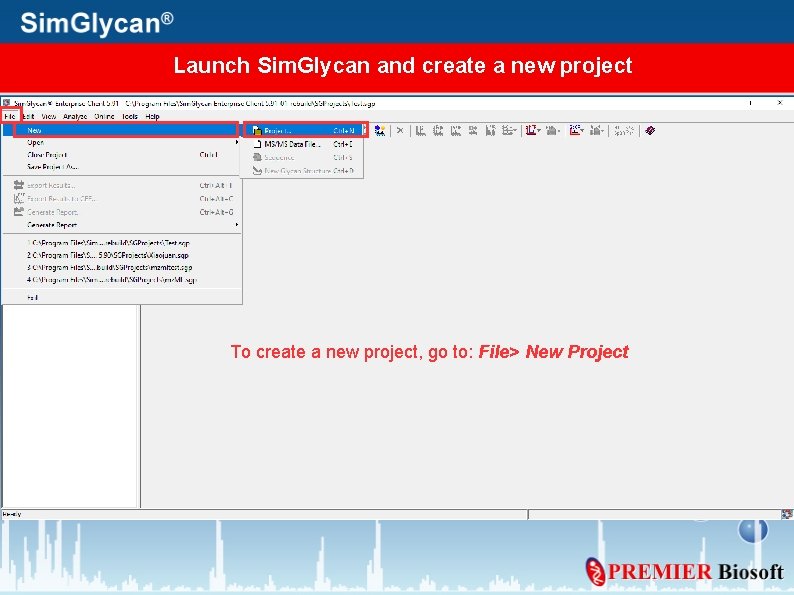
Launch Sim. Glycan and create a new project To create a new project, go to: File> New Project
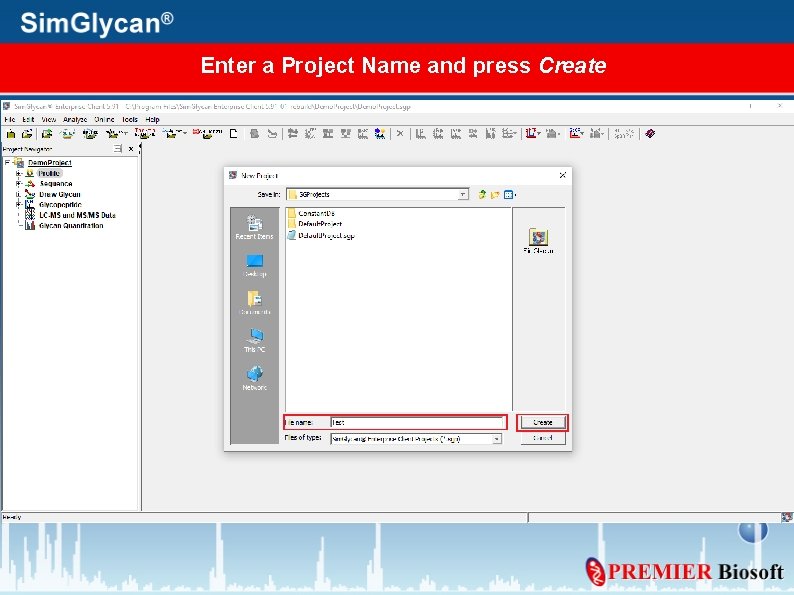
Enter a Project Name and press Create
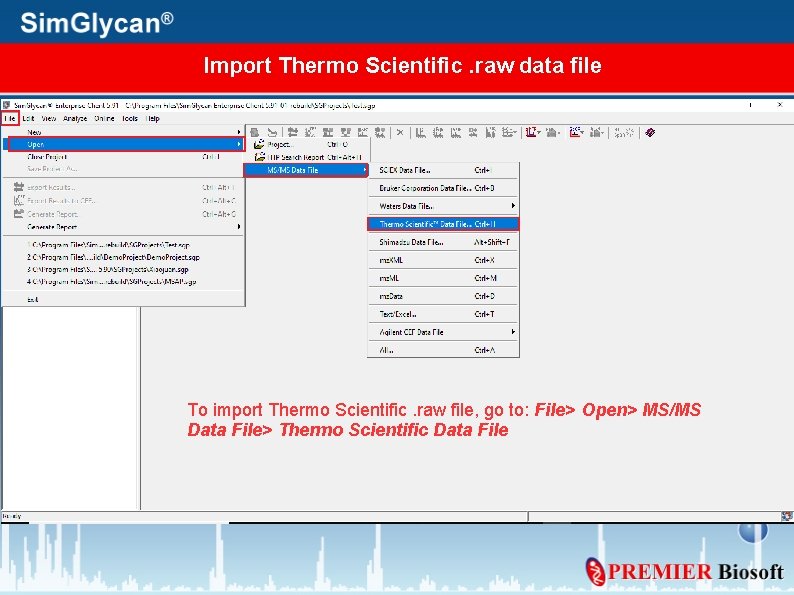
Import Thermo Scientific. raw data file To import Thermo Scientific. raw file, go to: File> Open> MS/MS Data File> Thermo Scientific Data File
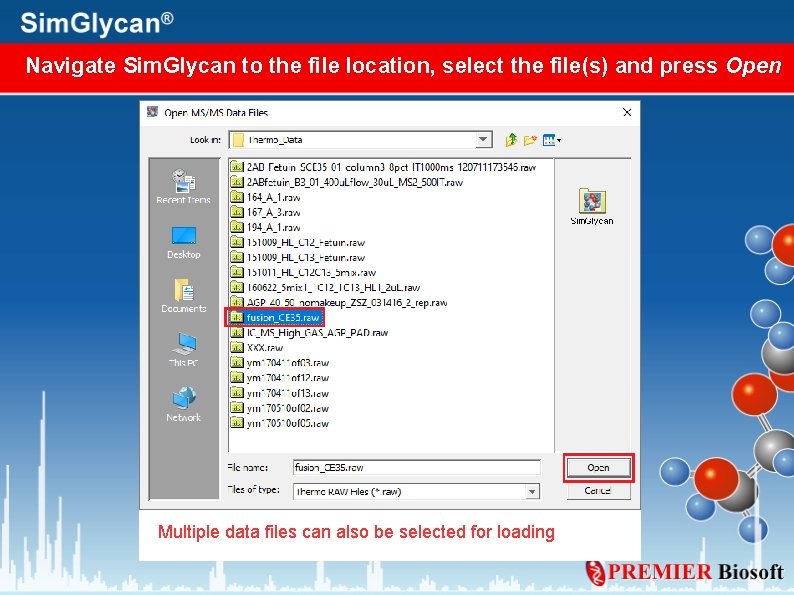
Navigate Sim. Glycan to the file location, select the file(s) and press Open Multiple data files can also be selected for loading
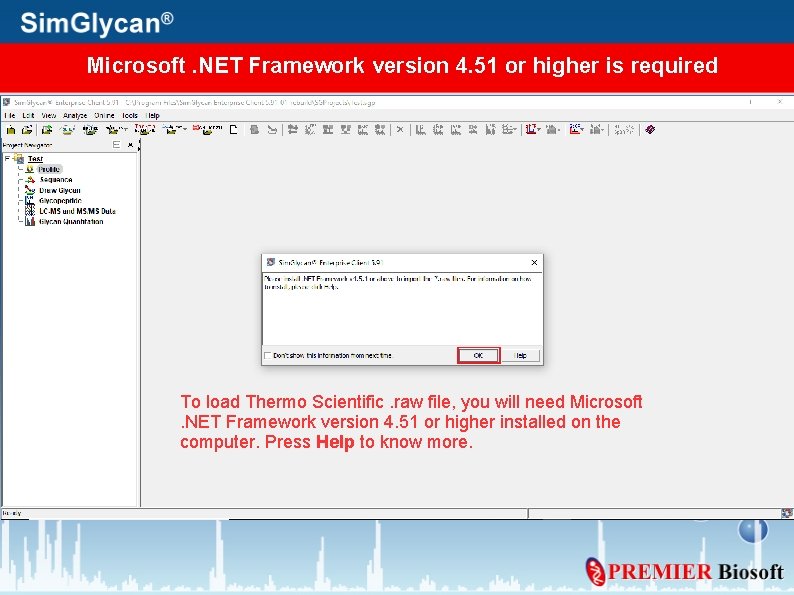
Microsoft. NET Framework version 4. 51 or higher is required To load Thermo Scientific. raw file, you will need Microsoft. NET Framework version 4. 51 or higher installed on the computer. Press Help to know more.
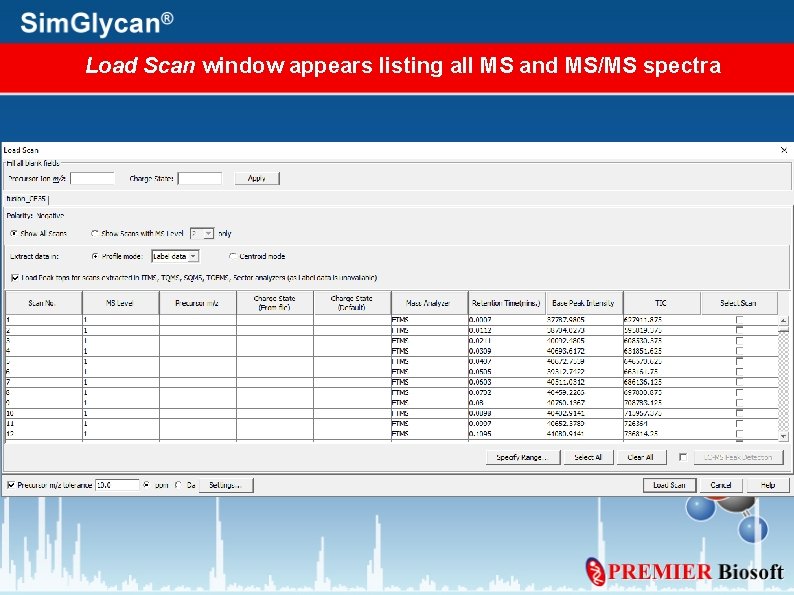
Load Scan window appears listing all MS and MS/MS spectra
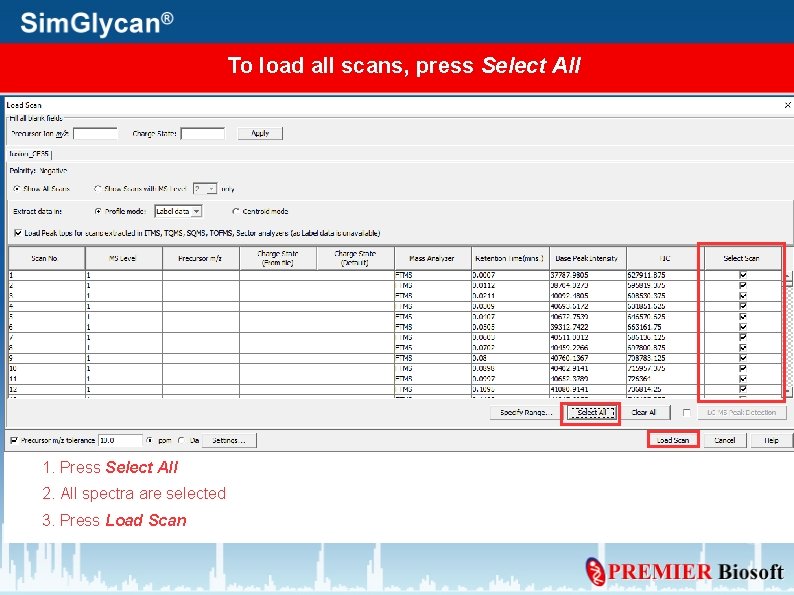
To load all scans, press Select All 1. Press Select All 2. All spectra are selected 3. Press Load Scan
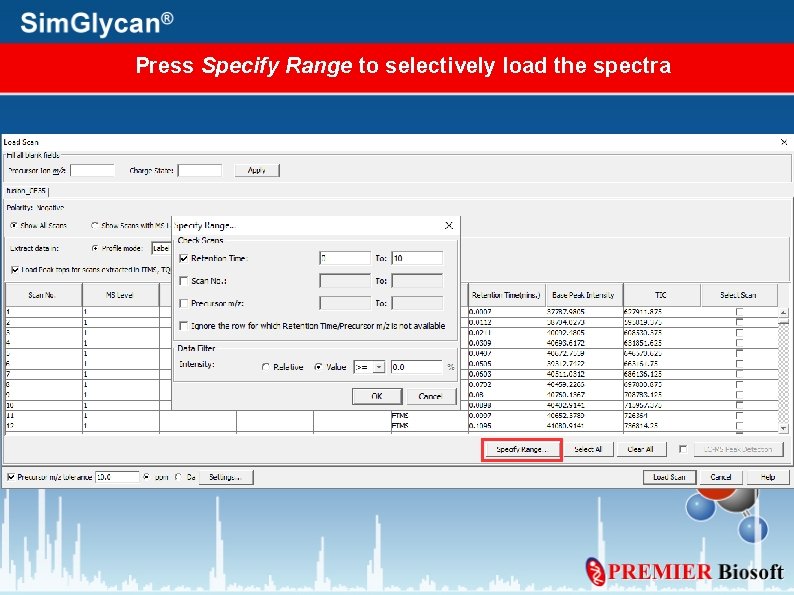
Press Specify Range to selectively load the spectra
Upon pressing Load Scan, data loading starts
Once completed, loaded data file appears under Profile node
Click on LC-MS and MS/MS Data node
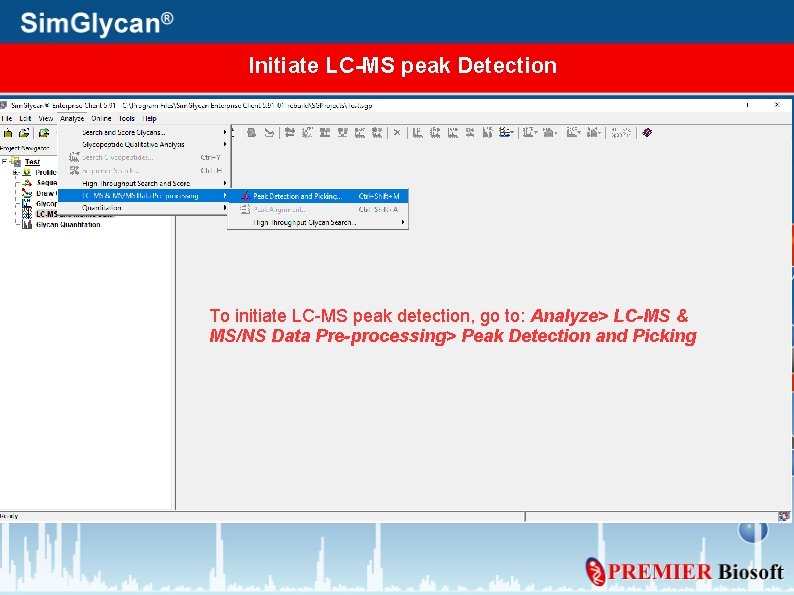
Initiate LC-MS peak Detection To initiate LC-MS peak detection, go to: Analyze> LC-MS & MS/NS Data Pre-processing> Peak Detection and Picking
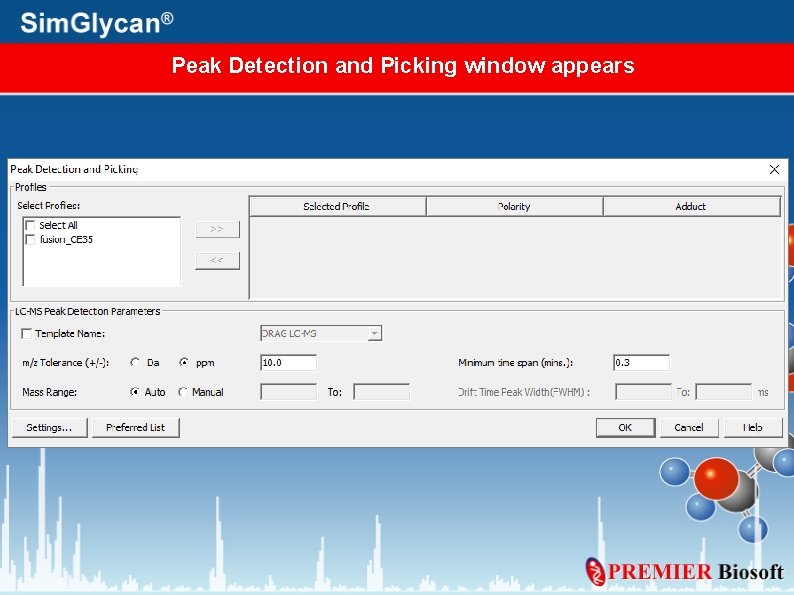
Peak Detection and Picking window appears
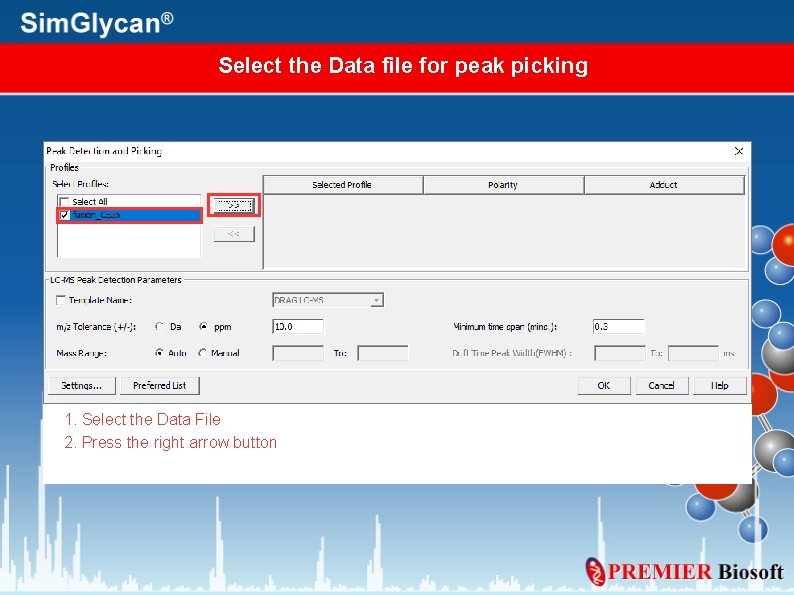
Select the Data file for peak picking 1. Select the Data File 2. Press the right arrow button
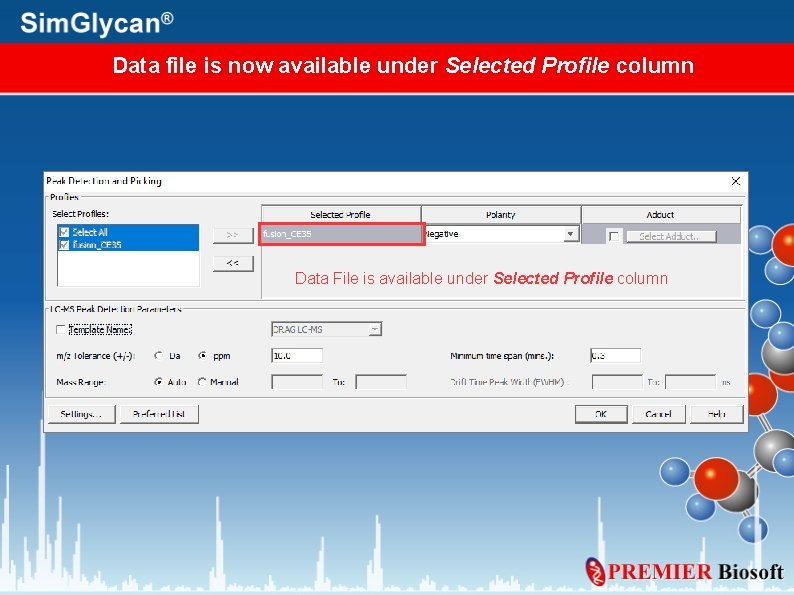
Data file is now available under Selected Profile column Data File is available under Selected Profile column
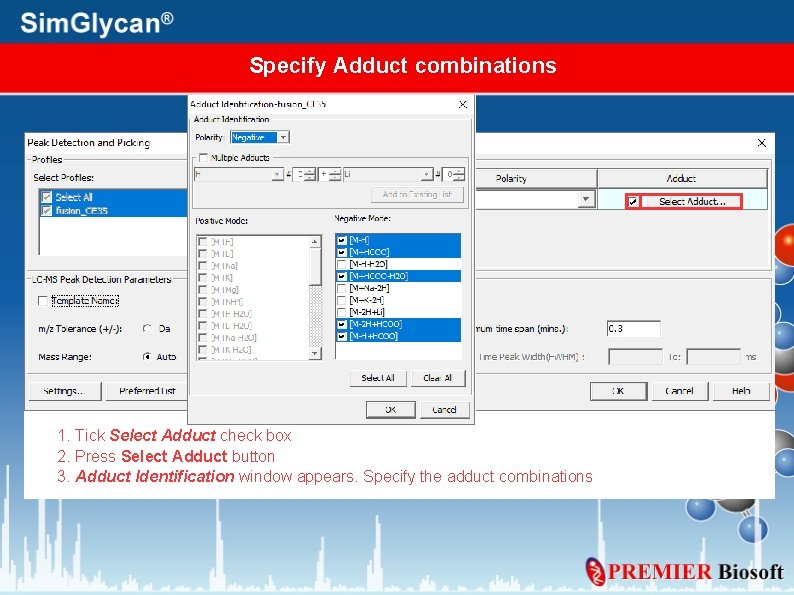
Specify Adduct combinations 1. Tick Select Adduct check box 2. Press Select Adduct button 3. Adduct Identification window appears. Specify the adduct combinations
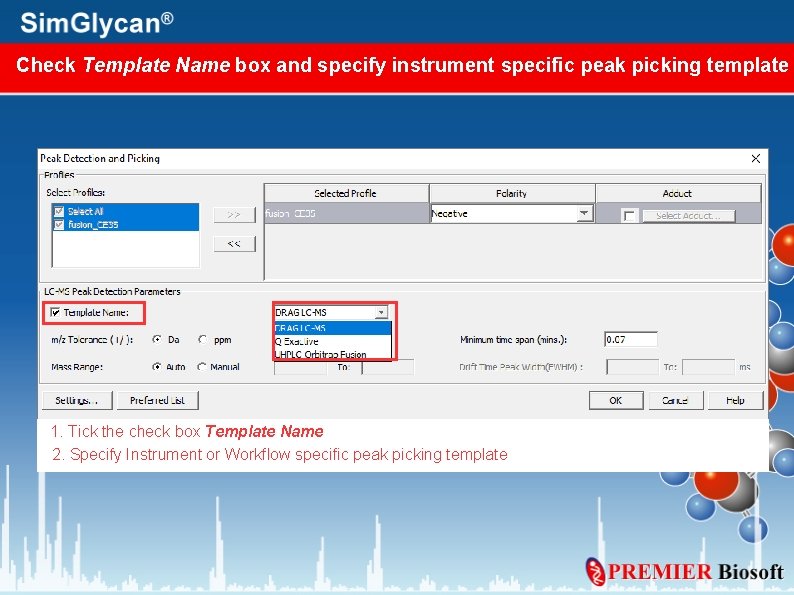
Check Template Name box and specify instrument specific peak picking template 1. Tick the check box Template Name 2. Specify Instrument or Workflow specific peak picking template
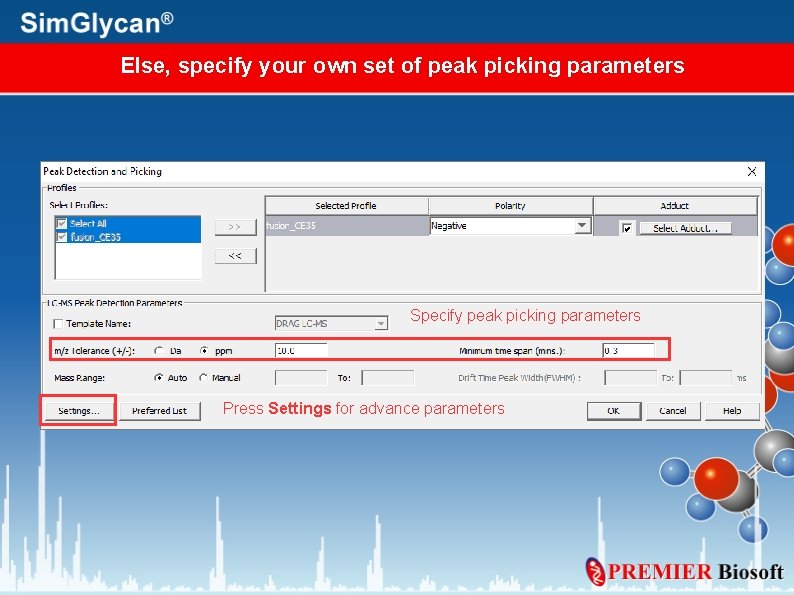
Else, specify your own set of peak picking parameters Specify peak picking parameters Press Settings for advance parameters
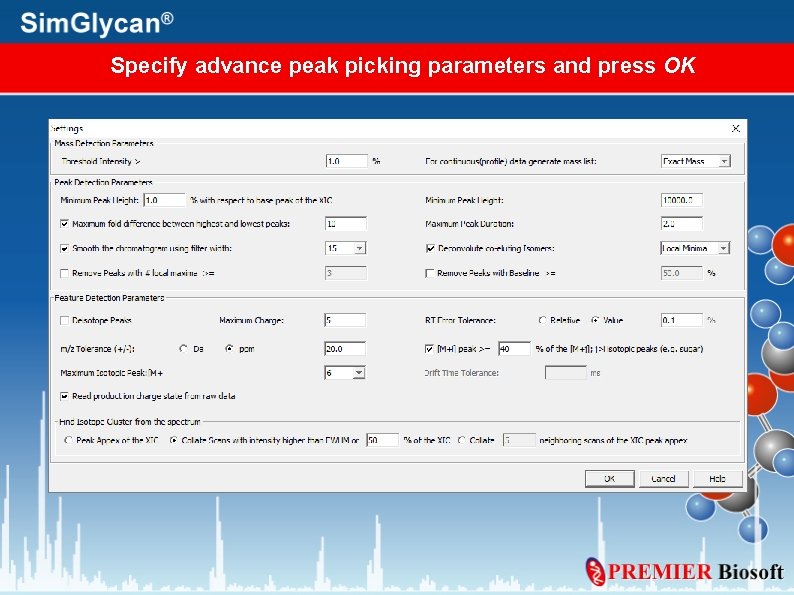
Specify advance peak picking parameters and press OK
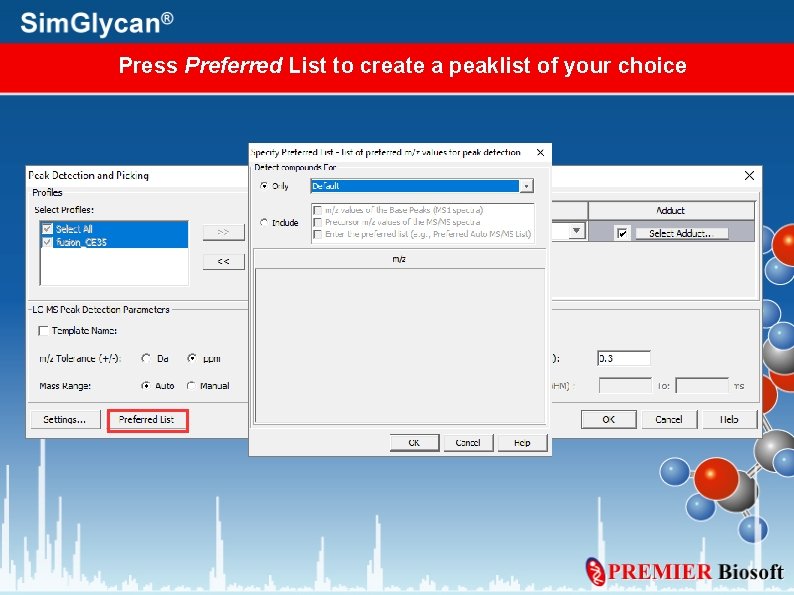
Press Preferred List to create a peaklist of your choice
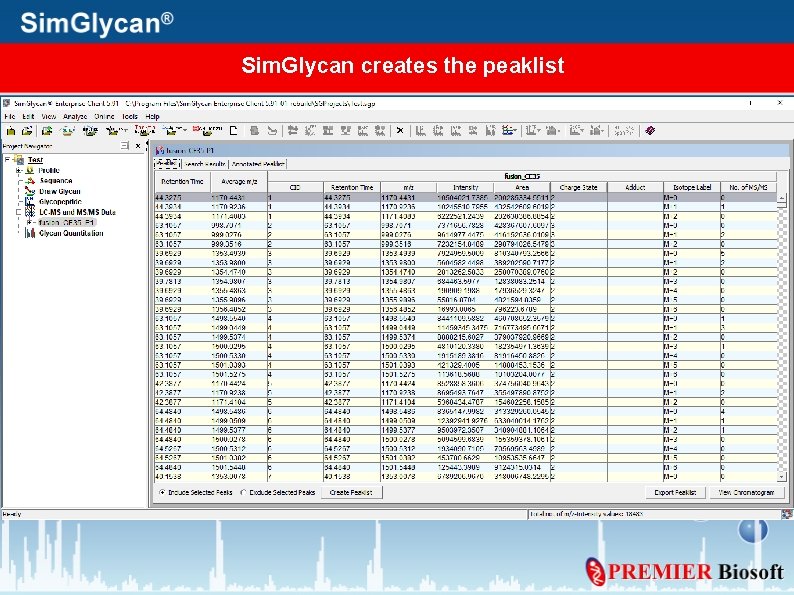
Sim. Glycan creates the peaklist
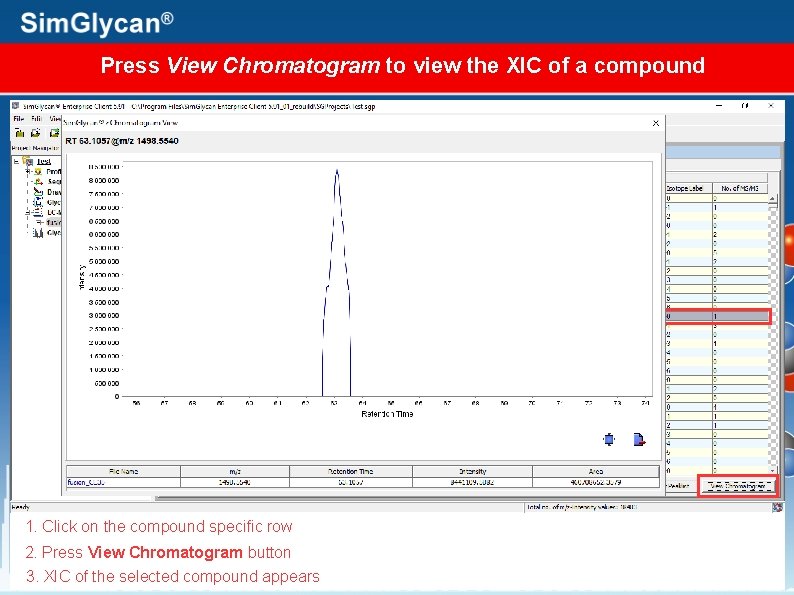
Press View Chromatogram to view the XIC of a compound 1. Click on the compound specific row 2. Press View Chromatogram button 3. XIC of the selected compound appears
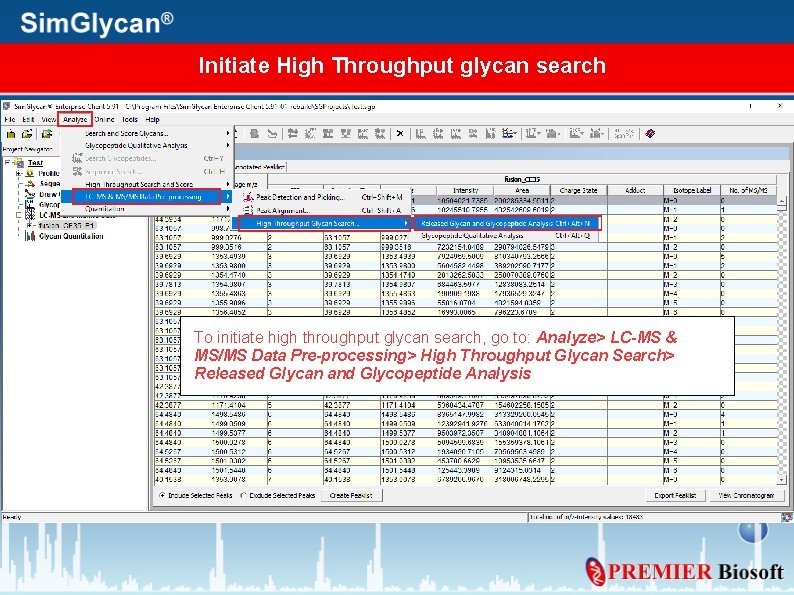
Initiate High Throughput glycan search To initiate high throughput glycan search, go to: Analyze> LC-MS & MS/MS Data Pre-processing> High Throughput Glycan Search> Released Glycan and Glycopeptide Analysis
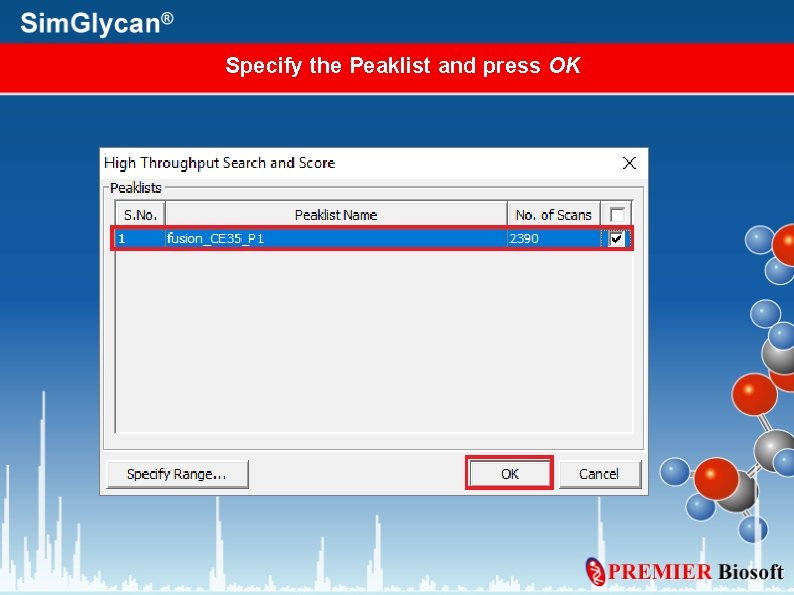
Specify the Peaklist and press OK
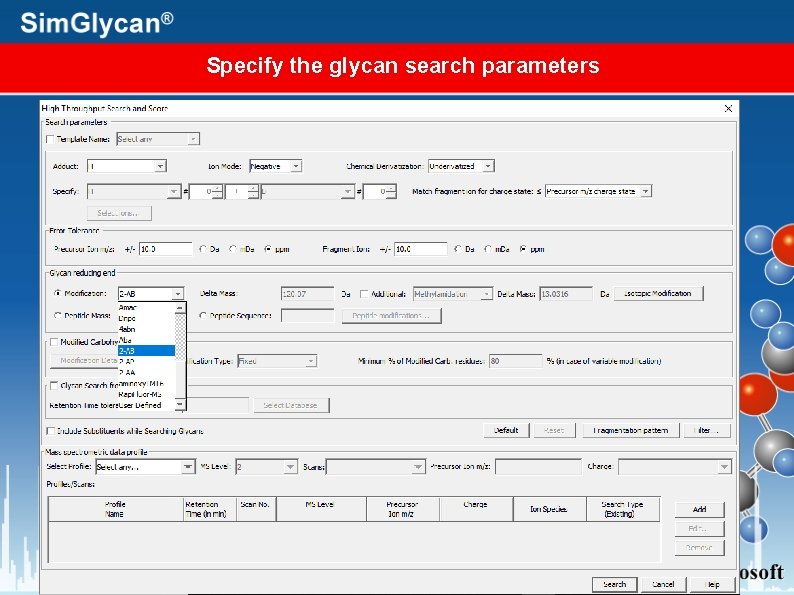
Specify the glycan search parameters
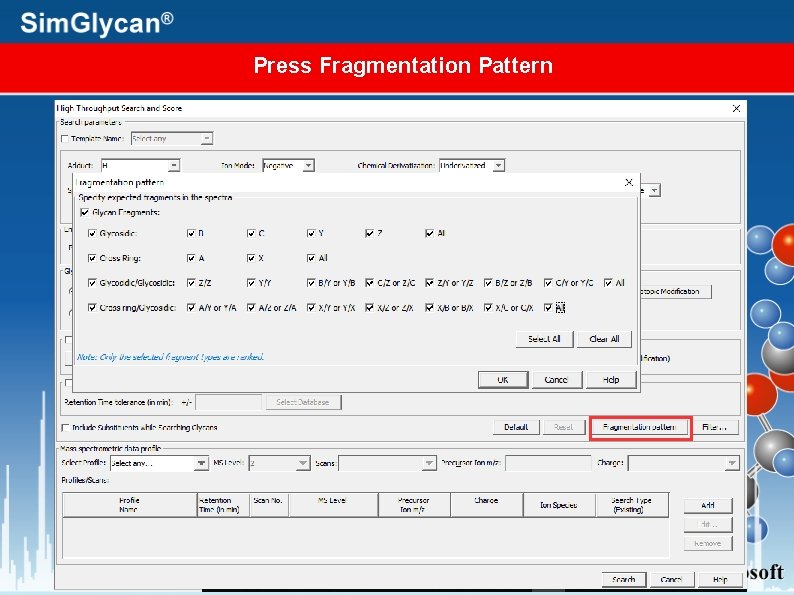
Press Fragmentation Pattern
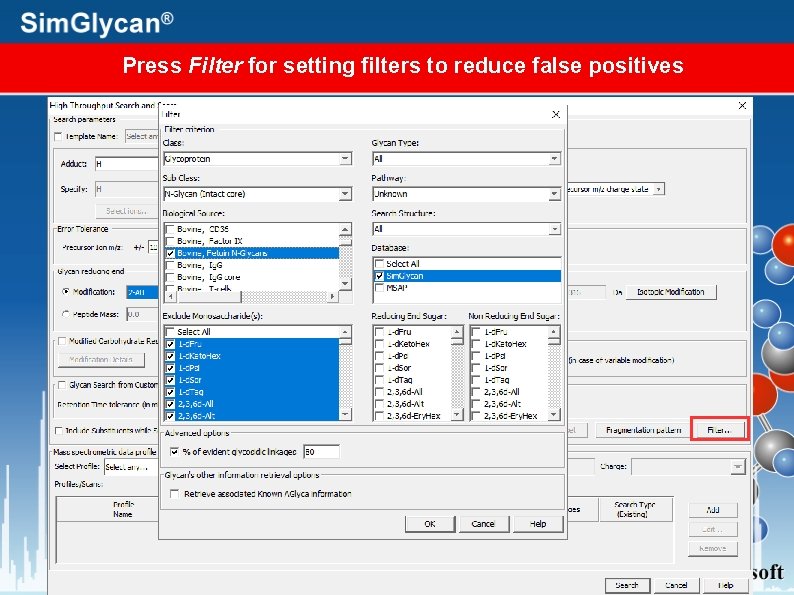
Press Filter for setting filters to reduce false positives
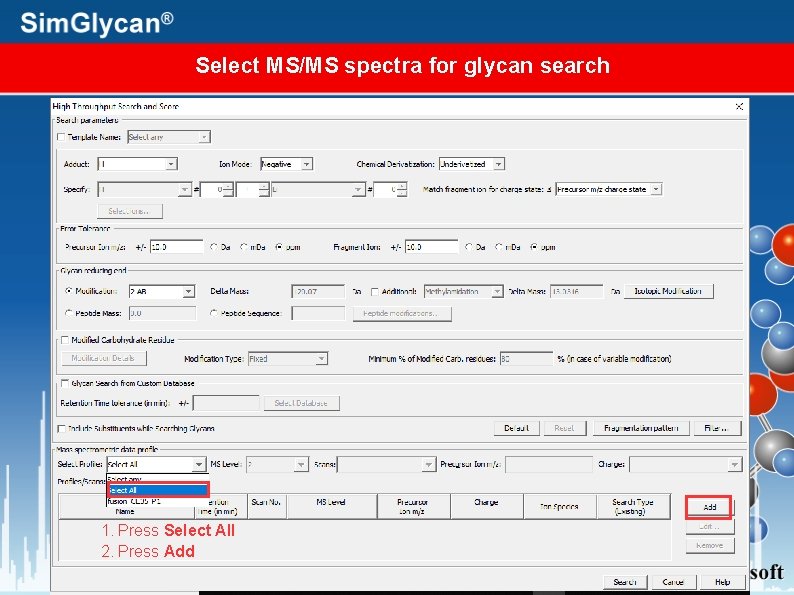
Select MS/MS spectra for glycan search 1. Press Select All 2. Press Add
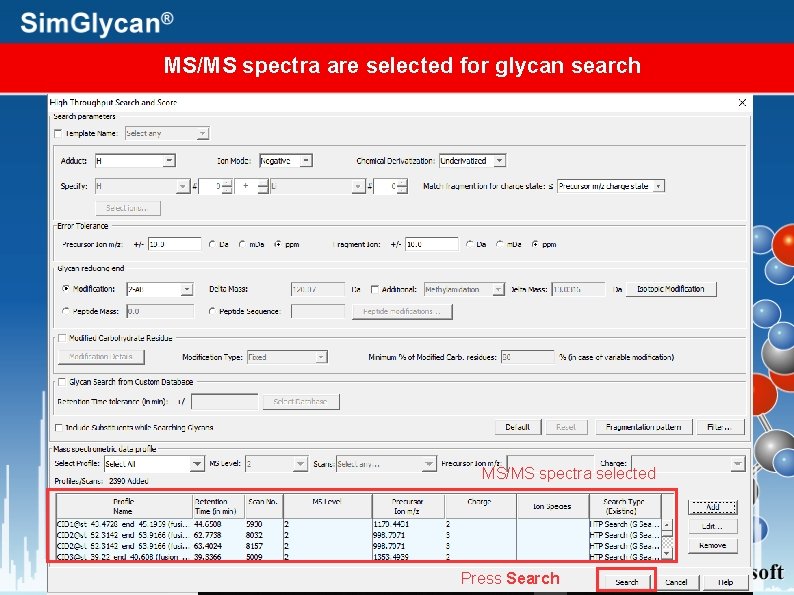
MS/MS spectra are selected for glycan search MS/MS spectra selected Press Search
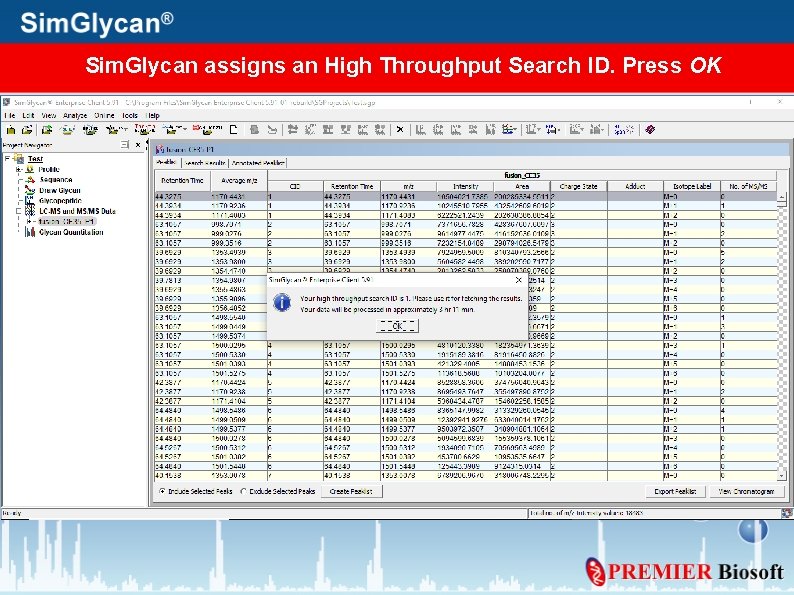
Sim. Glycan assigns an High Throughput Search ID. Press OK
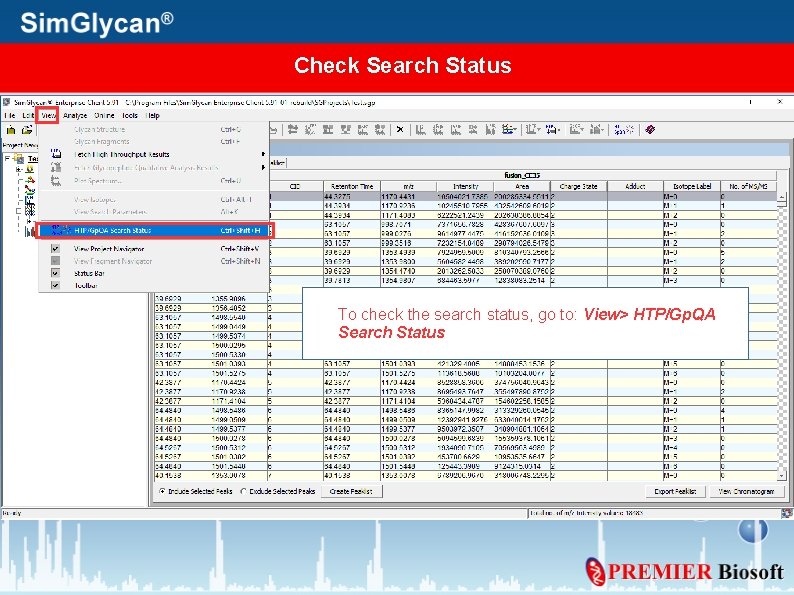
Check Search Status To check the search status, go to: View> HTP/Gp. QA Search Status
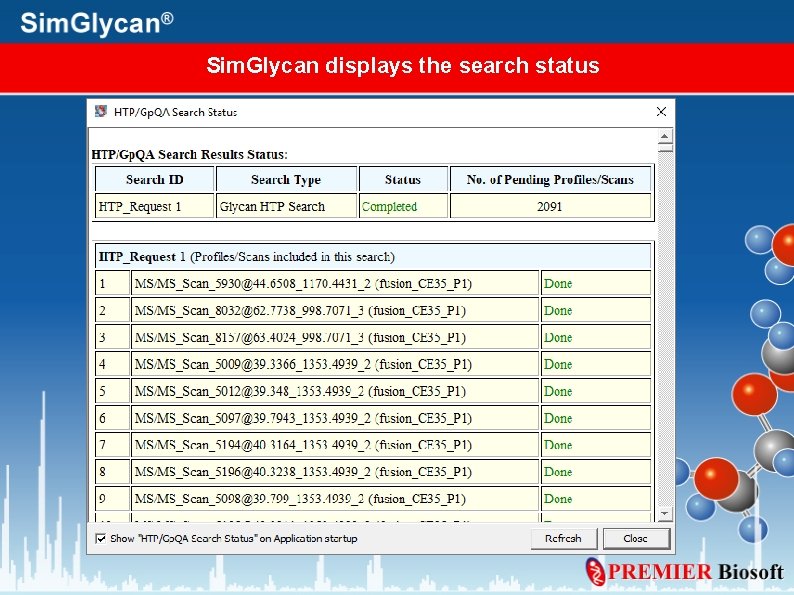
Sim. Glycan displays the search status
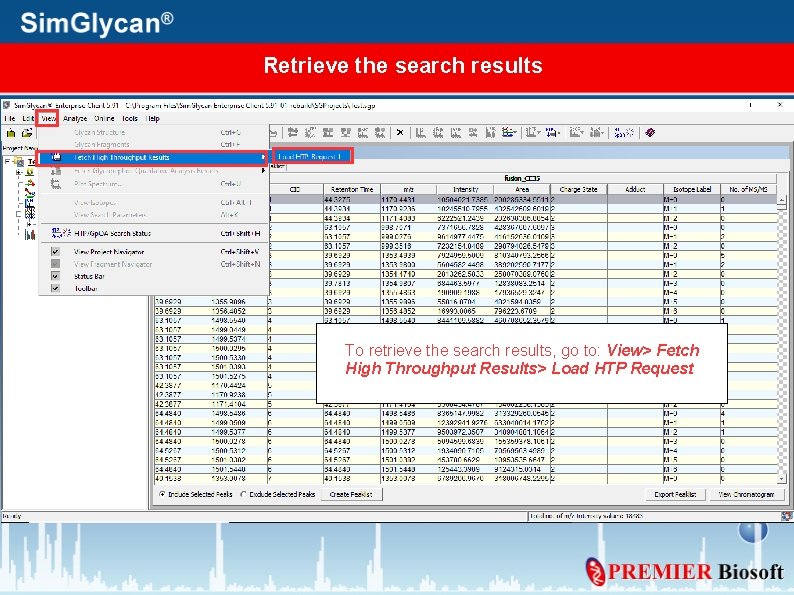
Retrieve the search results To retrieve the search results, go to: View> Fetch High Throughput Results> Load HTP Request
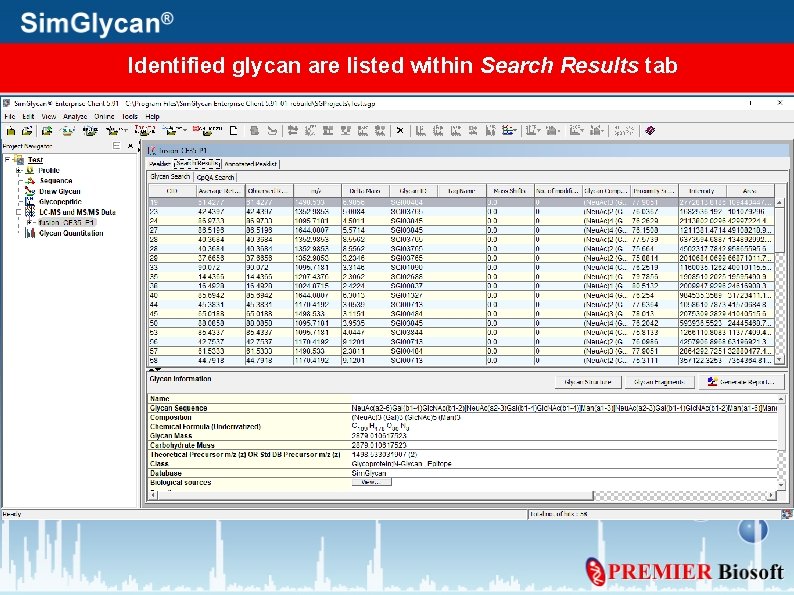
Identified glycan are listed within Search Results tab
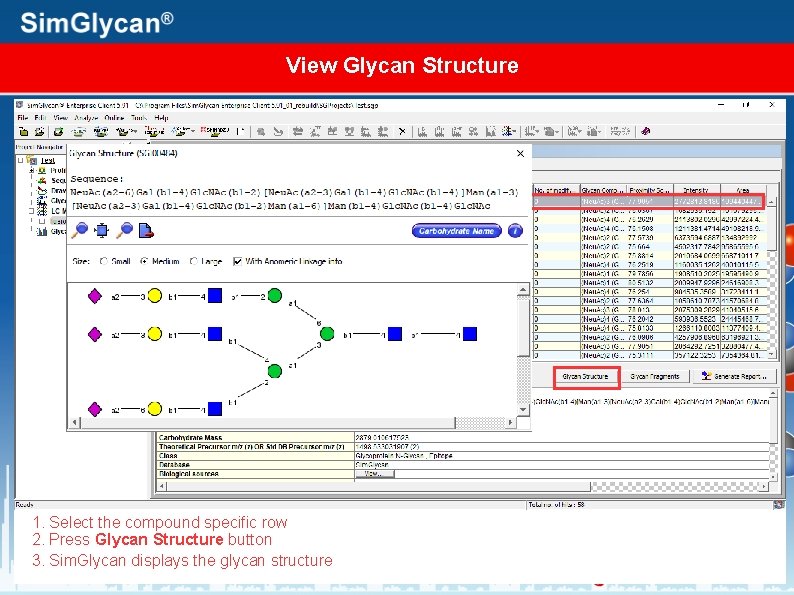
View Glycan Structure 1. Select the compound specific row 2. Press Glycan Structure button 3. Sim. Glycan displays the glycan structure
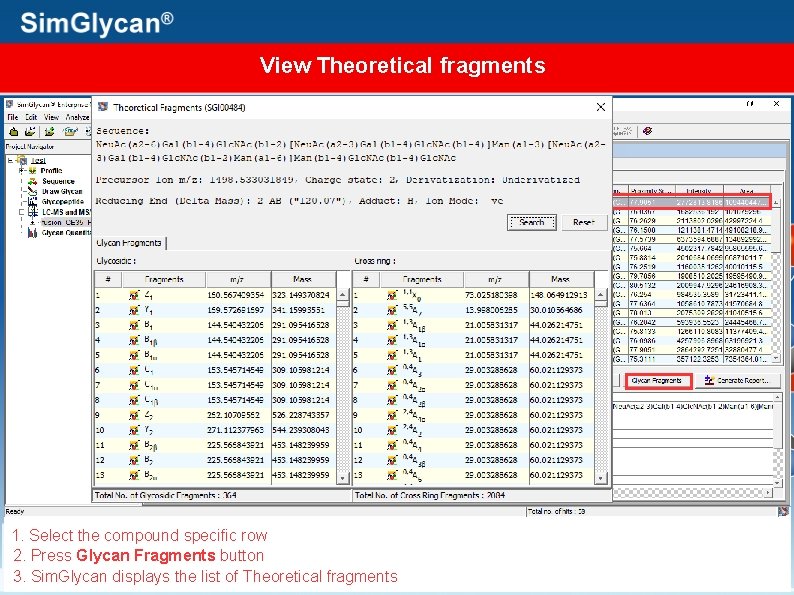
View Theoretical fragments 1. Select the compound specific row 2. Press Glycan Fragments button 3. Sim. Glycan displays the list of Theoretical fragments
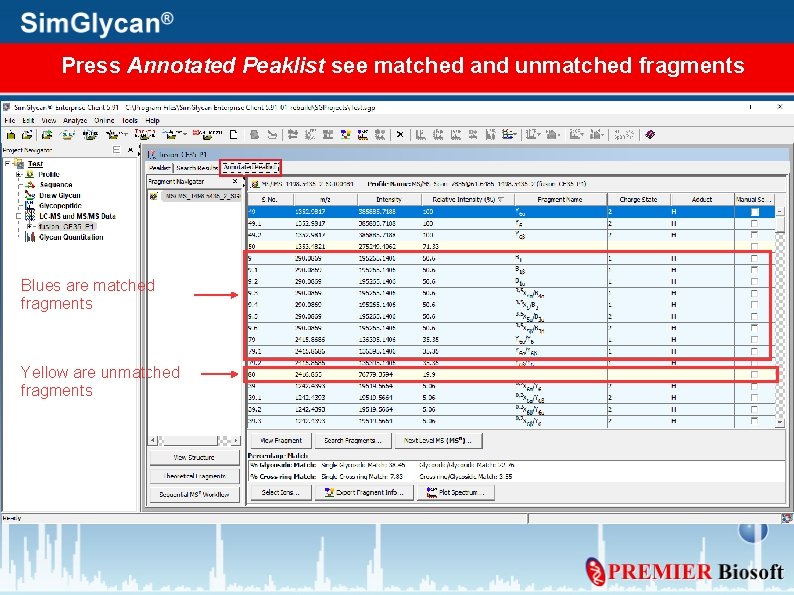
Press Annotated Peaklist see matched and unmatched fragments Blues are matched fragments Yellow are unmatched fragments
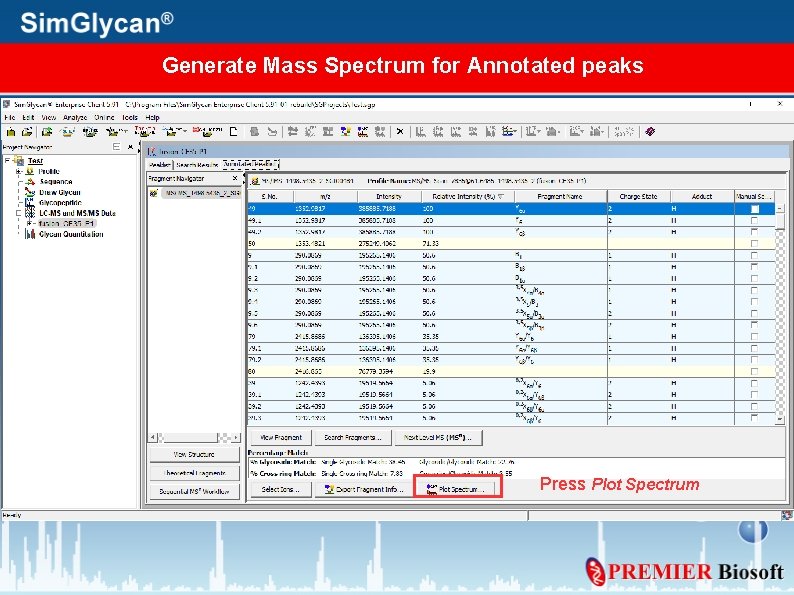
Generate Mass Spectrum for Annotated peaks Press Plot Spectrum
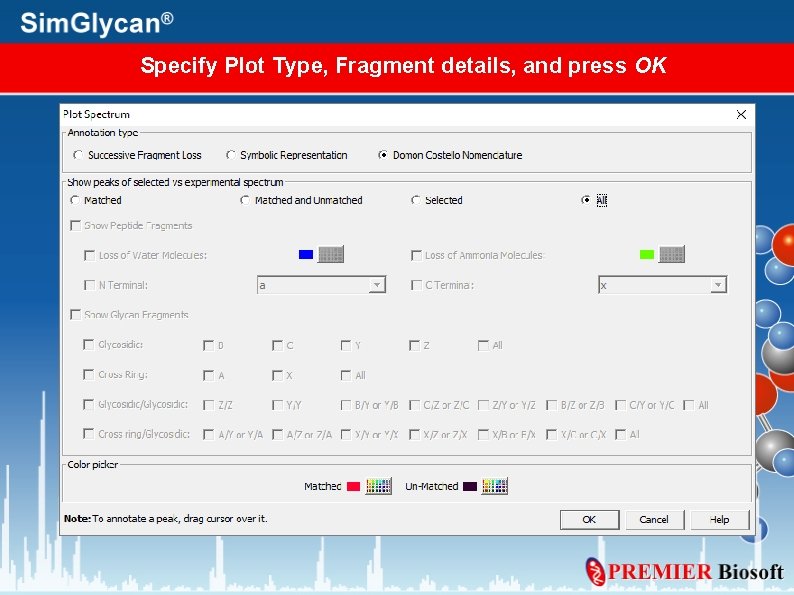
Specify Plot Type, Fragment details, and press OK
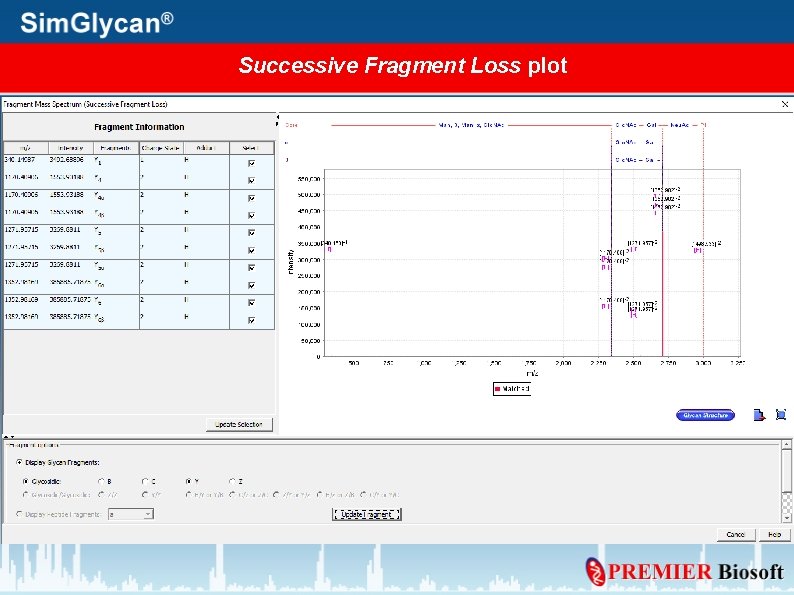
Successive Fragment Loss plot
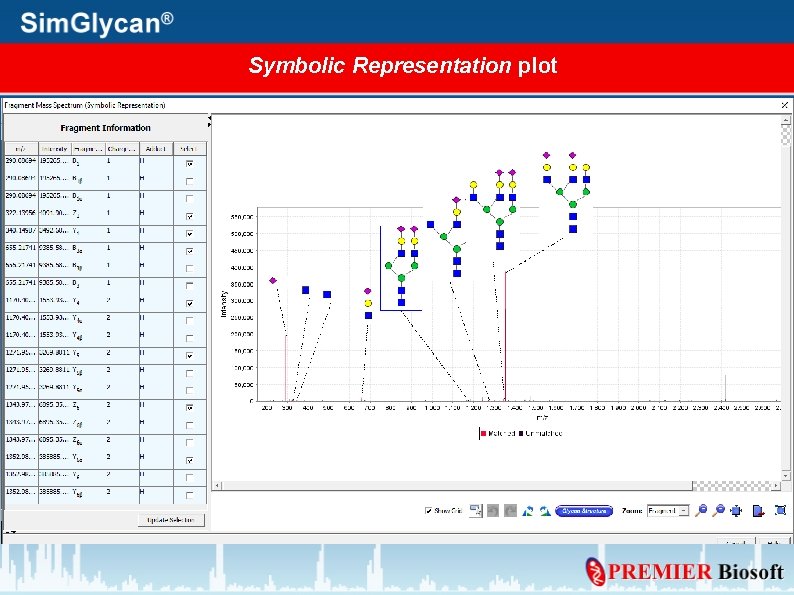
Symbolic Representation plot
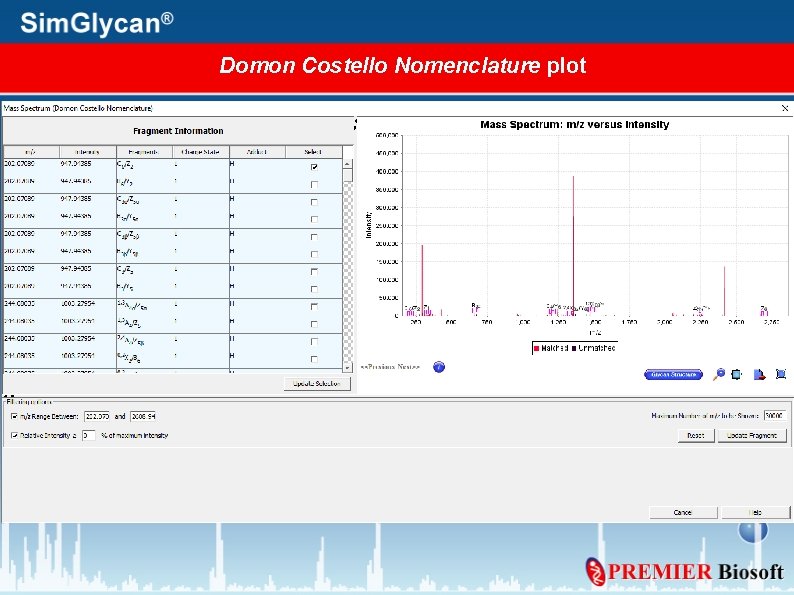
Domon Costello Nomenclature plot
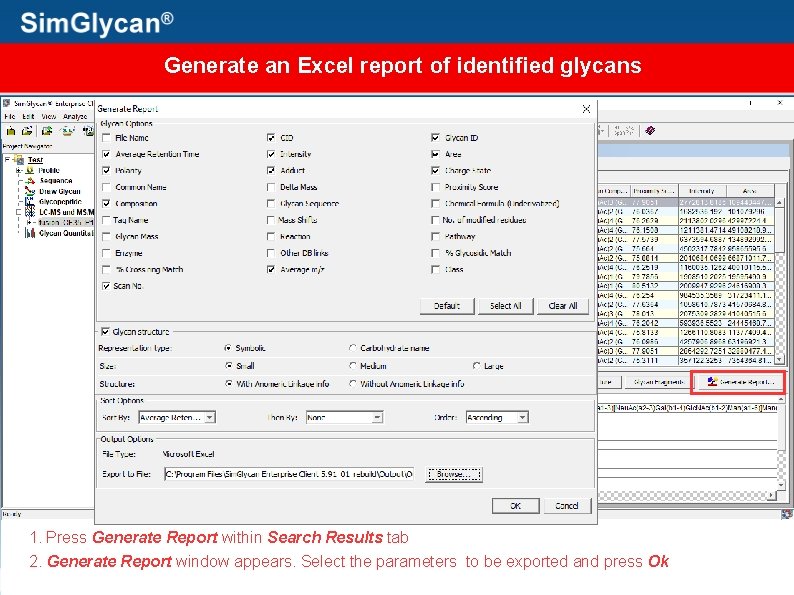
Generate an Excel report of identified glycans 1. Press Generate Report within Search Results tab 2. Generate Report window appears. Select the parameters to be exported and press Ok
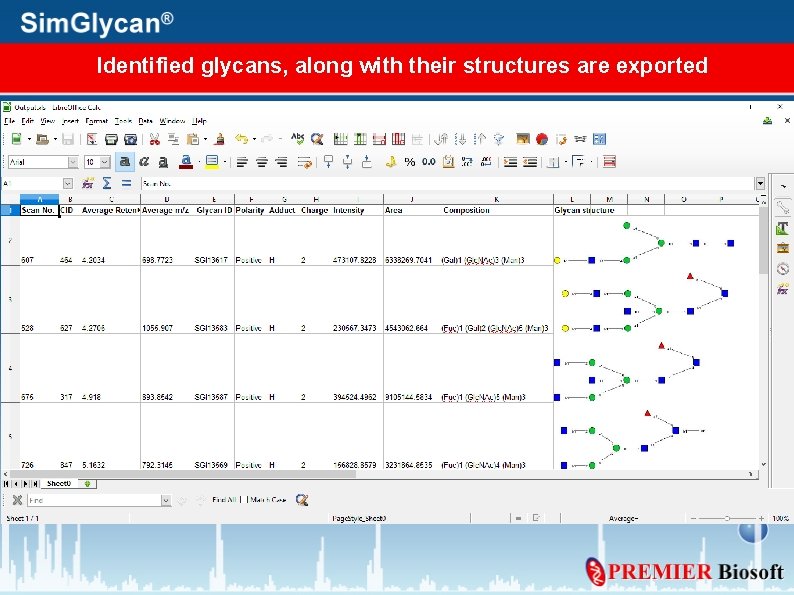
Identified glycans, along with their structures are exported

Thank You!
- Slides: 48