Predicting protein structure and function Protein function GenomeDNA







- Slides: 15

Predicting protein structure and function

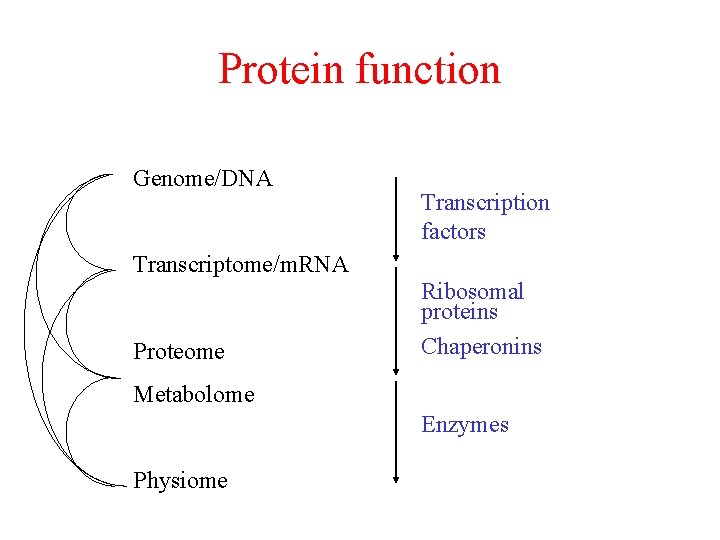
Protein function Genome/DNA Transcription factors Transcriptome/m. RNA Proteome Ribosomal proteins Chaperonins Metabolome Enzymes Physiome

Protein function Not all proteins are enzymes: -crystallin: eye lens protein – needs to stay stable and transparent for a lifetime (very little turnover in the eye lens)

What can happen to protein function through evolution Proteins can have multiple functions (and sometimes many -- Ig). Enzyme function is defined by specificity and activity Through evolution: • Function and specificity can stay the same • Function stays same but specificity changes • Change to some similar function (e. g. somewhere else in metabolic system) • Change to completely new function

How to arrive at a given function • Divergent evolution – homologous proteins –proteins have same structure and “sameish” function • Convergent evolution – analogous proteins – different structure but same function • Question: can homologous proteins change structure (and function)?

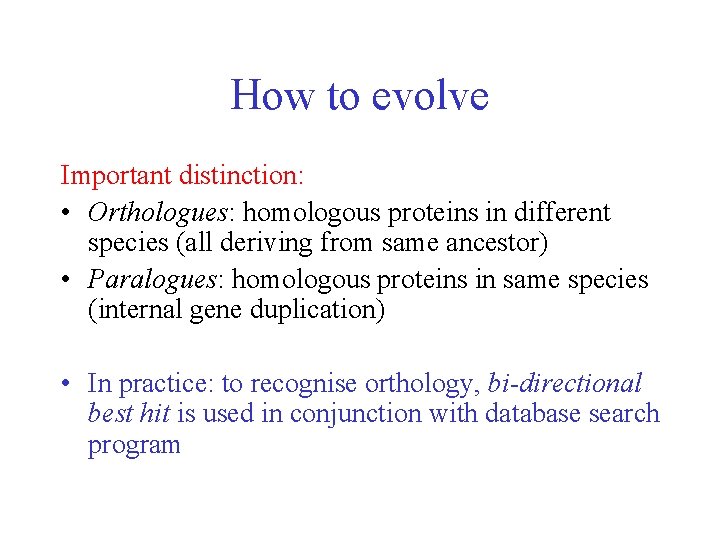
How to evolve Important distinction: • Orthologues: homologous proteins in different species (all deriving from same ancestor) • Paralogues: homologous proteins in same species (internal gene duplication) • In practice: to recognise orthology, bi-directional best hit is used in conjunction with database search program

How to evolve By addition of domains (at either end of protein sequence) – Lesk book page 108 Often through gene duplication followed by divergence

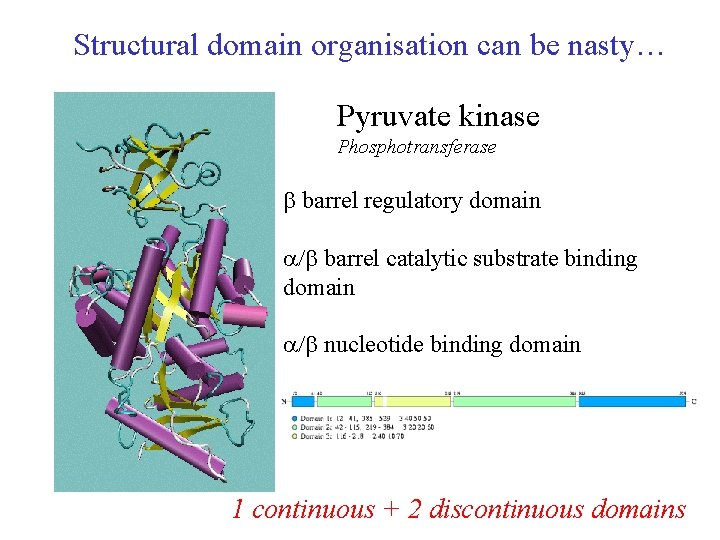
Structural domain organisation can be nasty… Pyruvate kinase Phosphotransferase barrel regulatory domain / barrel catalytic substrate binding domain / nucleotide binding domain 1 continuous + 2 discontinuous domains

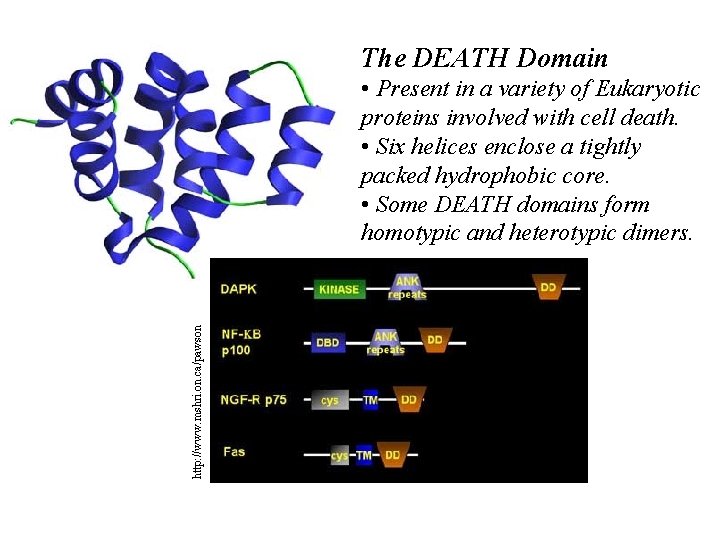
The DEATH Domain http: //www. mshri. on. ca/pawson • Present in a variety of Eukaryotic proteins involved with cell death. • Six helices enclose a tightly packed hydrophobic core. • Some DEATH domains form homotypic and heterotypic dimers.

How to predict function • Sequence information: Homology searching • Sequence-structure information: Fold recognition (Threading) • Structure-structure information: structure superpositioning

Flavodoxin fold

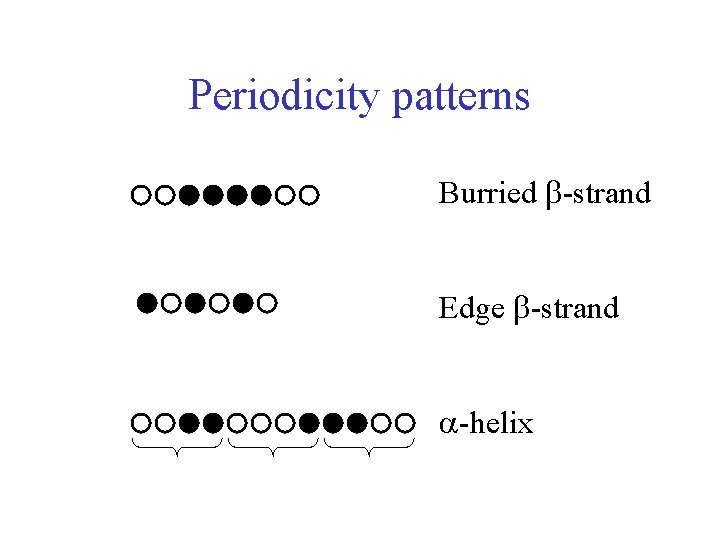
Rules of thumb when looking at a multiple alignment (MA) • • Hydrophobic residues are internal Gly (Thr, Ser) in loops MA: hydrophobic block -> internal -strand MA: alternating (1 -1) hydrophobic/hydrophilic => edge -strand • MA: alternating 2 -2 (or 3 -1) periodicity => -helix • MA: gaps in loops • MA: Conserved column => functional? => active site

Rules of thumb when looking at a multiple alignment (MA) • Active site residues are together in 3 D structure • Helices often cover up core of strands • Helices less extended than strands => more residues to cross protein • - - motif is right-handed in >95% of cases (with parallel strands) • MA: ‘inconsistent’ alignment columns and match errors! • Secondary structures have local anomalies, e. g. -bulges

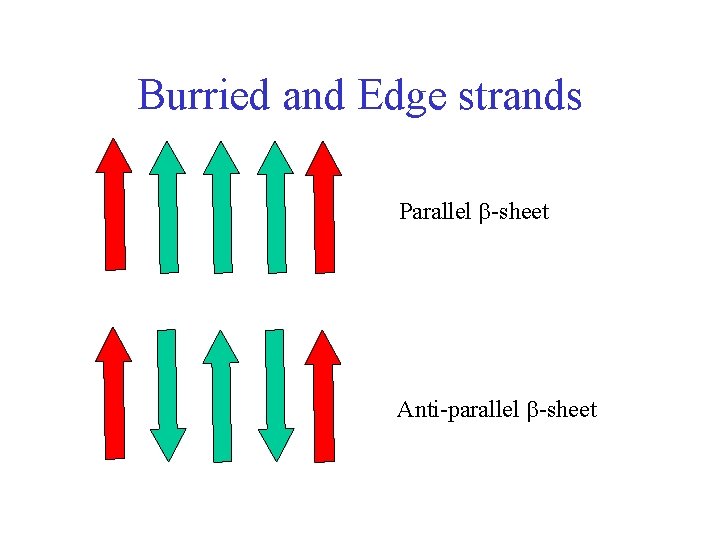
Burried and Edge strands Parallel -sheet Anti-parallel -sheet

Periodicity patterns Burried -strand Edge -strand -helix