Positional information fields boundaries and gradients Positional information

Positional information: fields, boundaries, and gradients Positional information is specified by (1) subdivision of larger fields of cells into smaller fields, and (2) specifying the "address" of each cell within the field. This is a recursive process that requires translation of gradients of gene expression into sharp boundaries, and initiation of new gradients by these boundaries What mechanisms are responsible for these transitions?
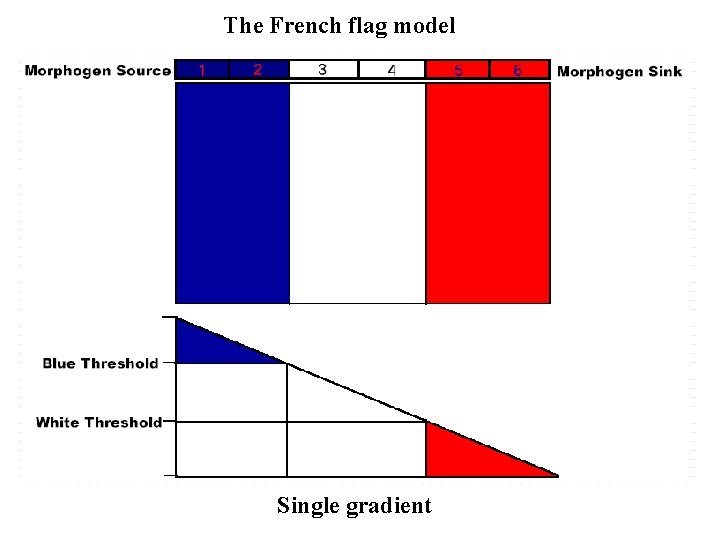
The French flag model Single gradient
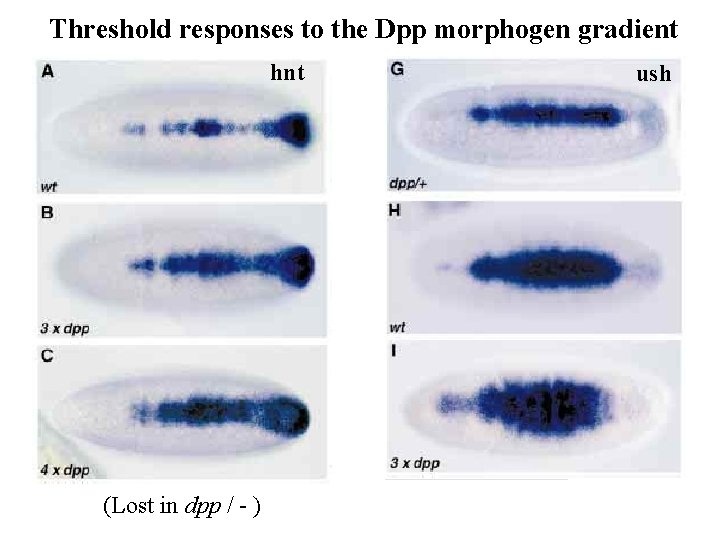
Threshold responses to the Dpp morphogen gradient hnt (Lost in dpp / - ) ush
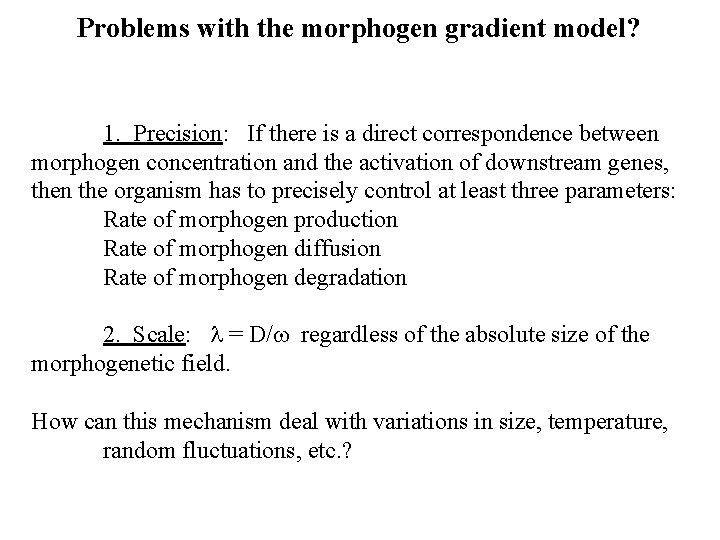
Problems with the morphogen gradient model? 1. Precision: If there is a direct correspondence between morphogen concentration and the activation of downstream genes, then the organism has to precisely control at least three parameters: Rate of morphogen production Rate of morphogen diffusion Rate of morphogen degradation 2. Scale: l = D/w regardless of the absolute size of the morphogenetic field. How can this mechanism deal with variations in size, temperature, random fluctuations, etc. ?
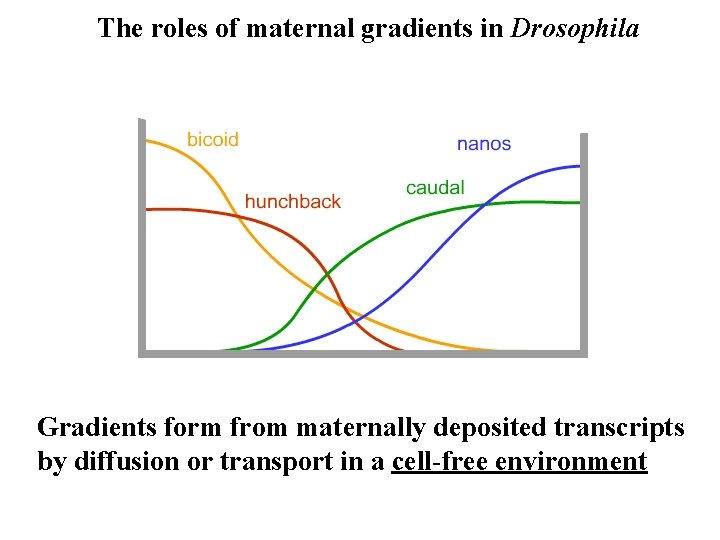
The roles of maternal gradients in Drosophila Gradients form from maternally deposited transcripts by diffusion or transport in a cell-free environment
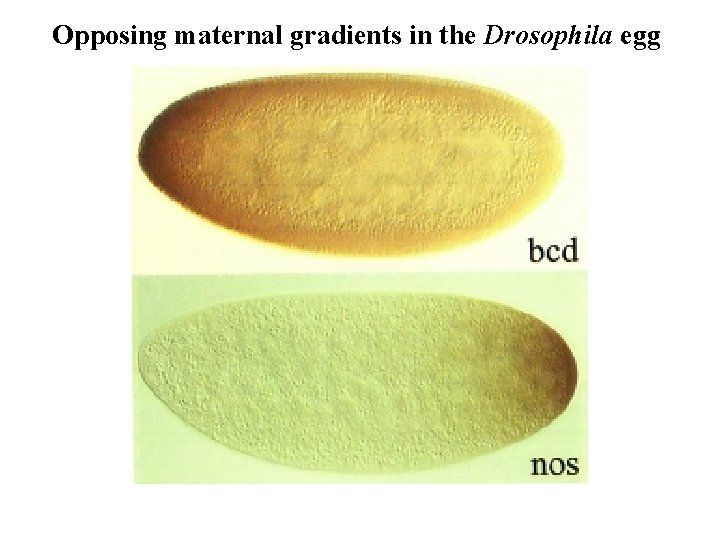
Opposing maternal gradients in the Drosophila egg
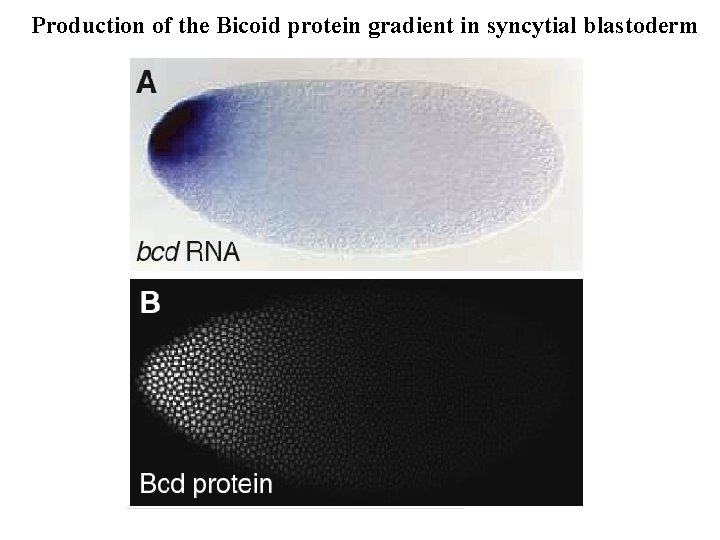
Production of the Bicoid protein gradient in syncytial blastoderm
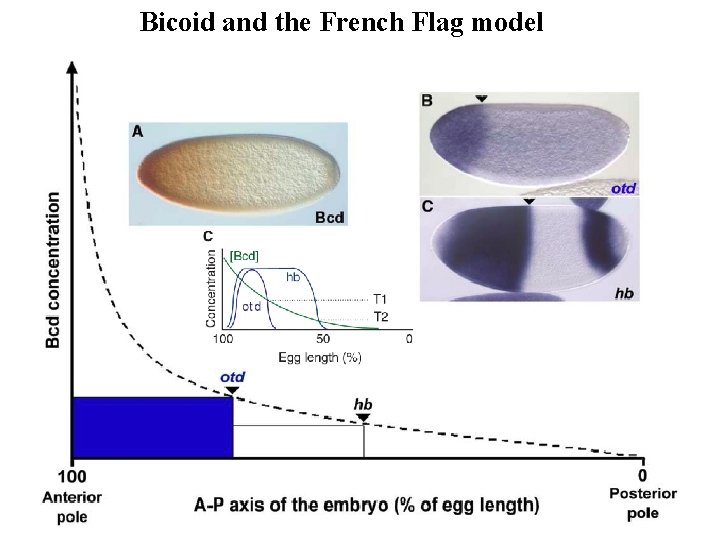
Bicoid and the French Flag model
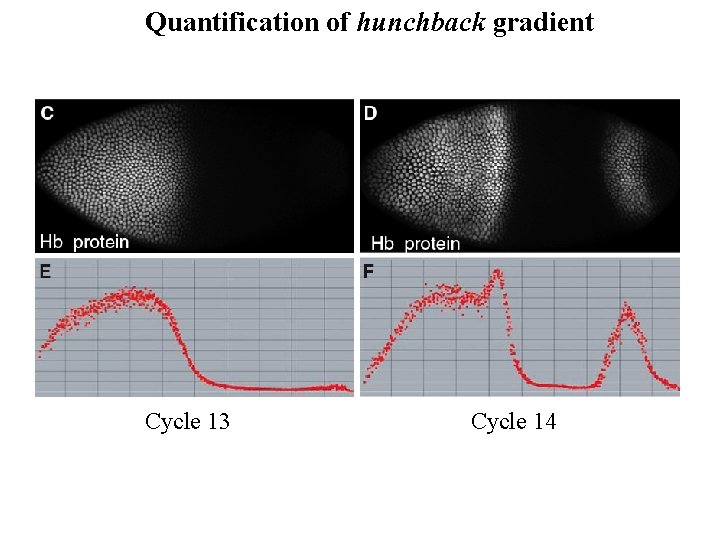
Quantification of hunchback gradient Cycle 13 Cycle 14
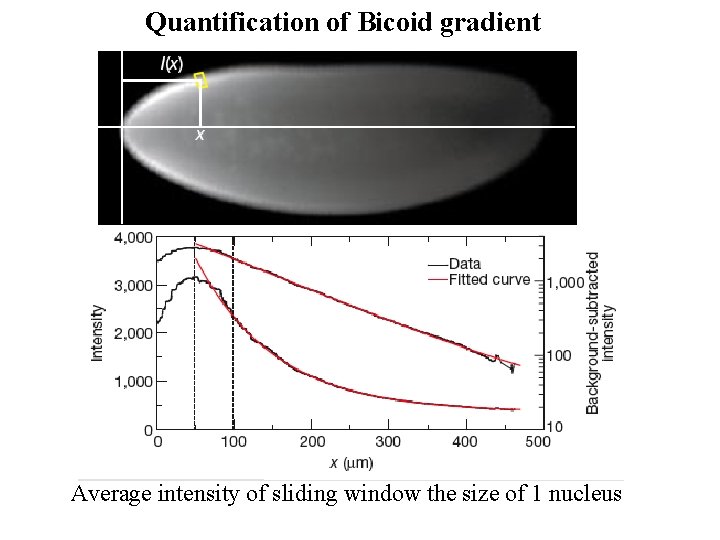
Quantification of Bicoid gradient Average intensity of sliding window the size of 1 nucleus
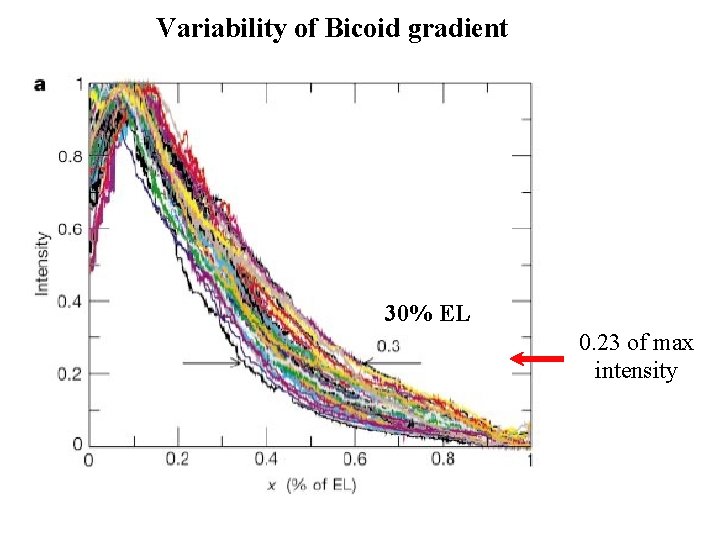
Variability of Bicoid gradient 30% EL 0. 23 of max intensity
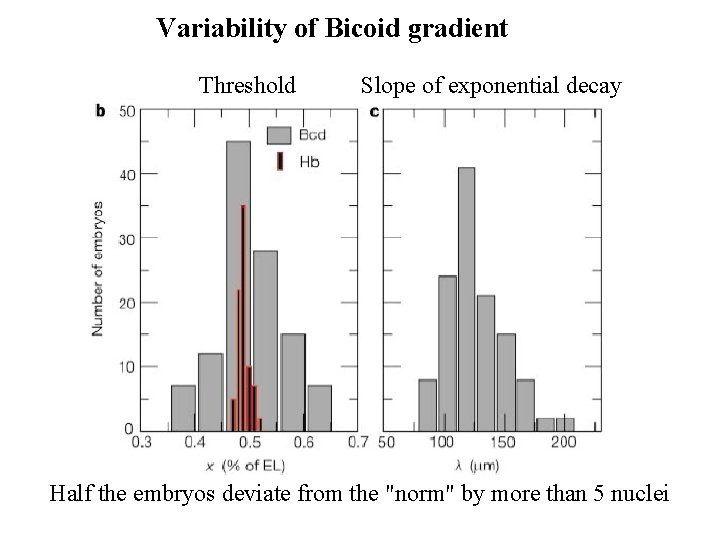
Variability of Bicoid gradient Threshold Slope of exponential decay Half the embryos deviate from the "norm" by more than 5 nuclei
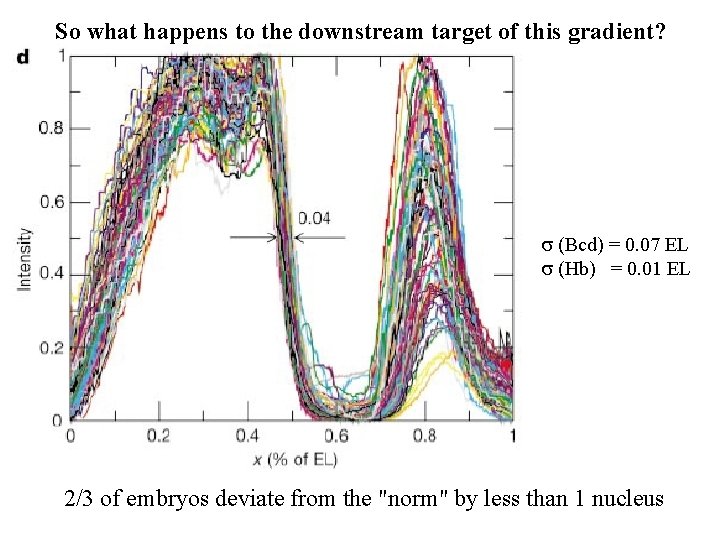
So what happens to the downstream target of this gradient? s (Bcd) = 0. 07 EL s (Hb) = 0. 01 EL 2/3 of embryos deviate from the "norm" by less than 1 nucleus
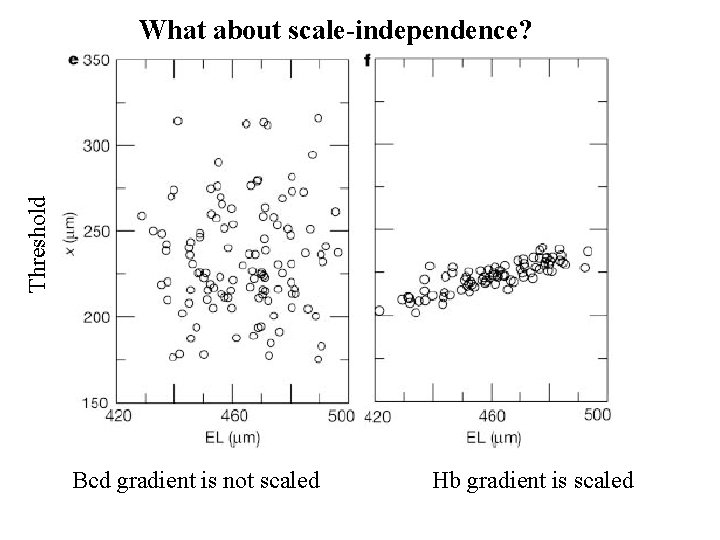
Threshold What about scale-independence? Bcd gradient is not scaled Hb gradient is scaled
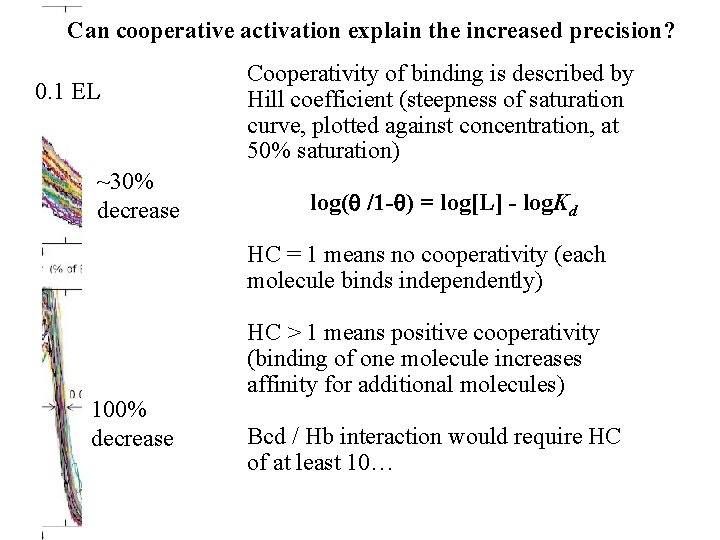
Can cooperative activation explain the increased precision? 0. 1 EL ~30% decrease Cooperativity of binding is described by Hill coefficient (steepness of saturation curve, plotted against concentration, at 50% saturation) log(q /1 -q) = log[L] - log. Kd HC = 1 means no cooperativity (each molecule binds independently) 100% decrease HC > 1 means positive cooperativity (binding of one molecule increases affinity for additional molecules) Bcd / Hb interaction would require HC of at least 10…
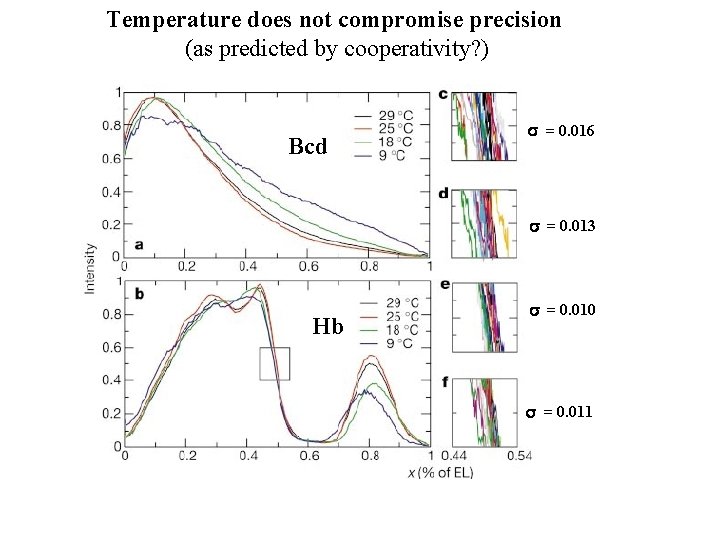
Temperature does not compromise precision (as predicted by cooperativity? ) Bcd s = 0. 016 s = 0. 013 Hb s = 0. 010 s = 0. 011
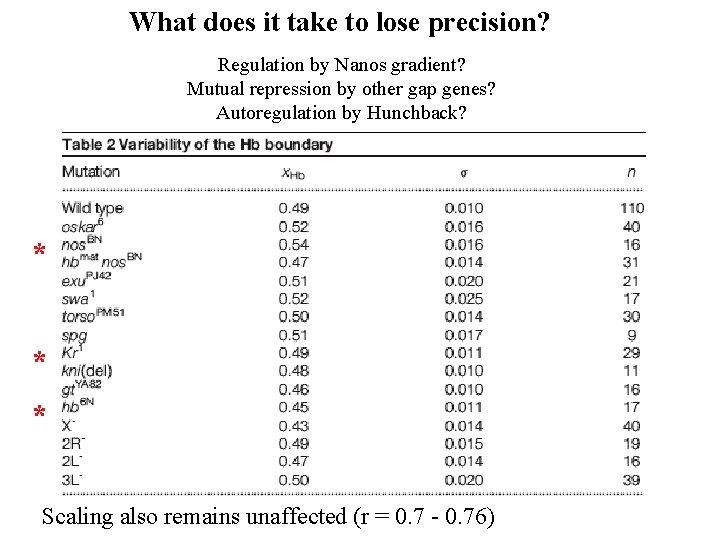
What does it take to lose precision? Regulation by Nanos gradient? Mutual repression by other gap genes? Autoregulation by Hunchback? * * * Scaling also remains unaffected (r = 0. 7 - 0. 76)
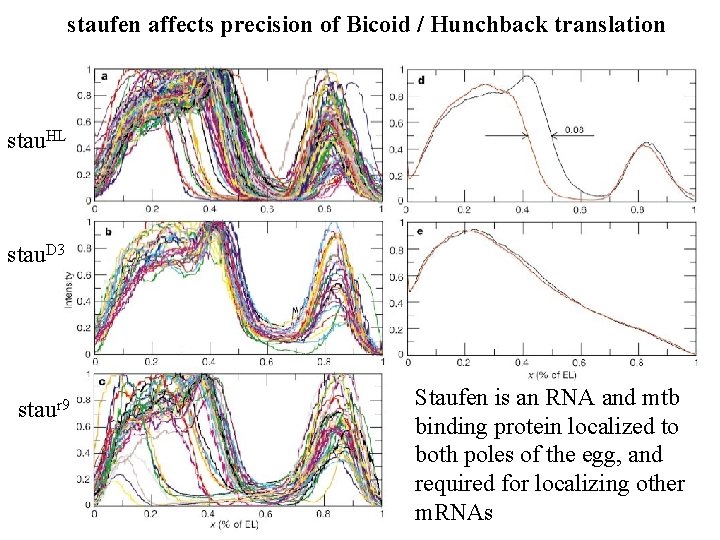
staufen affects precision of Bicoid / Hunchback translation stau. HL stau. D 3 staur 9 Staufen is an RNA and mtb binding protein localized to both poles of the egg, and required for localizing other m. RNAs
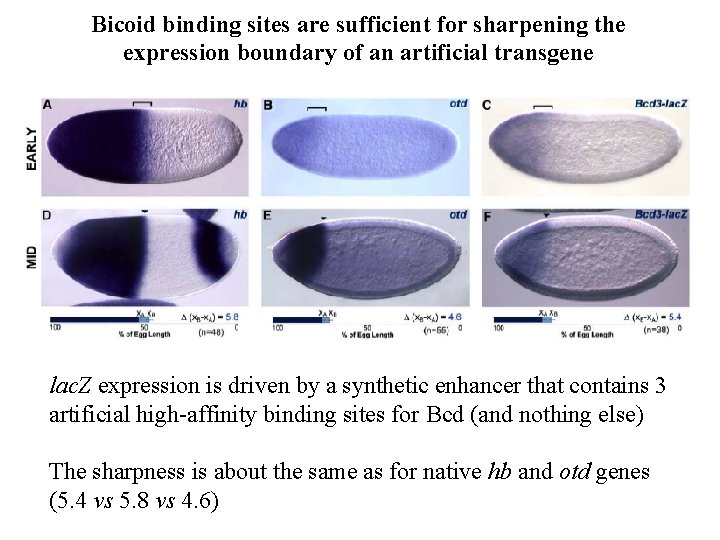
Bicoid binding sites are sufficient for sharpening the expression boundary of an artificial transgene lac. Z expression is driven by a synthetic enhancer that contains 3 artificial high-affinity binding sites for Bcd (and nothing else) The sharpness is about the same as for native hb and otd genes (5. 4 vs 5. 8 vs 4. 6)
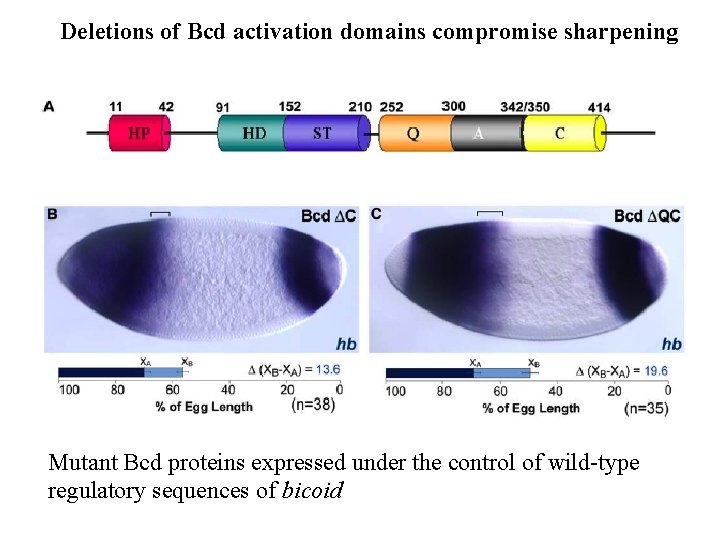
Deletions of Bcd activation domains compromise sharpening Mutant Bcd proteins expressed under the control of wild-type regulatory sequences of bicoid
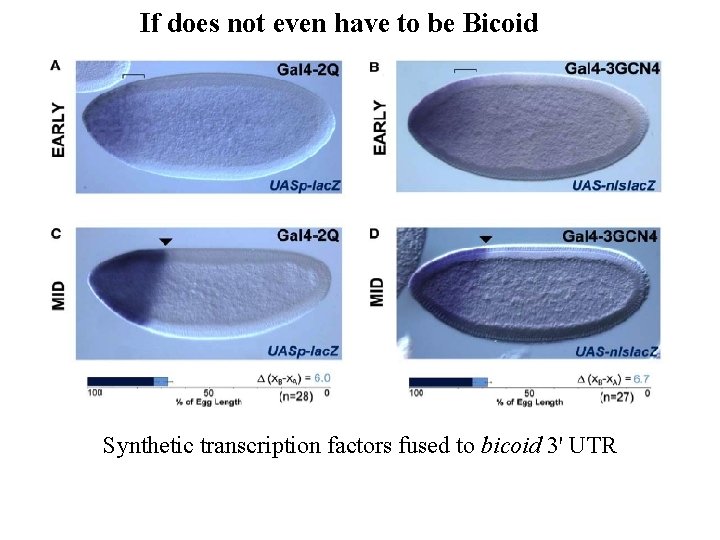
If does not even have to be Bicoid Synthetic transcription factors fused to bicoid 3' UTR
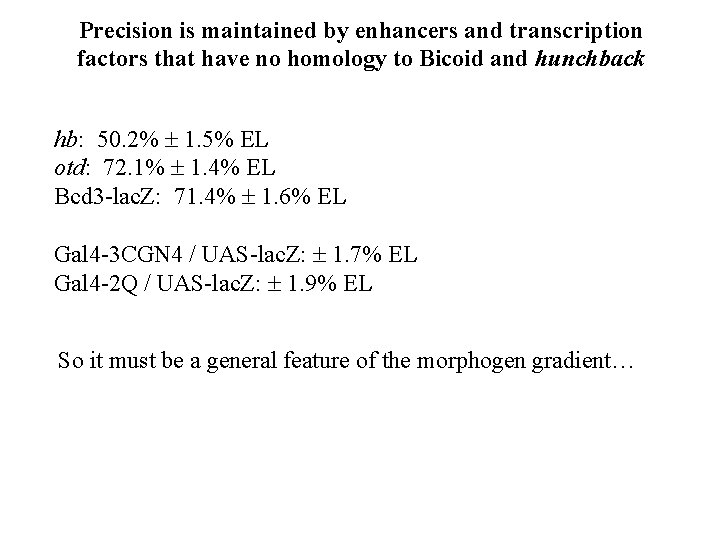
Precision is maintained by enhancers and transcription factors that have no homology to Bicoid and hunchback hb: 50. 2% 1. 5% EL otd: 72. 1% 1. 4% EL Bcd 3 -lac. Z: 71. 4% 1. 6% EL Gal 4 -3 CGN 4 / UAS-lac. Z: 1. 7% EL Gal 4 -2 Q / UAS-lac. Z: 1. 9% EL So it must be a general feature of the morphogen gradient…
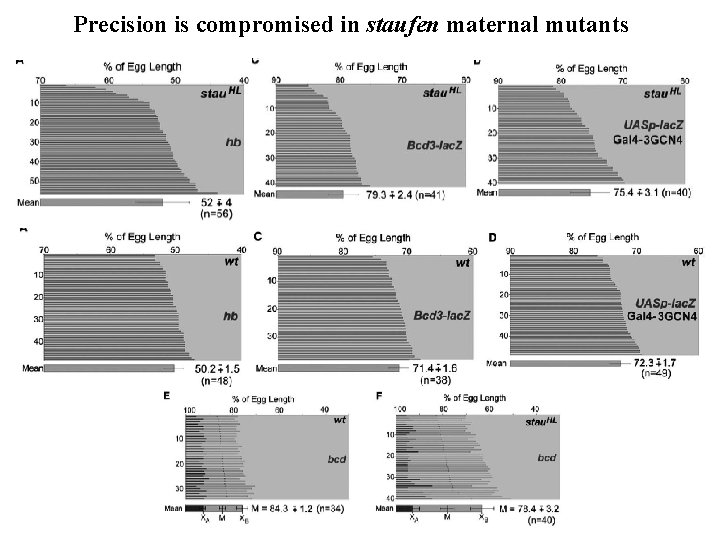
Precision is compromised in staufen maternal mutants
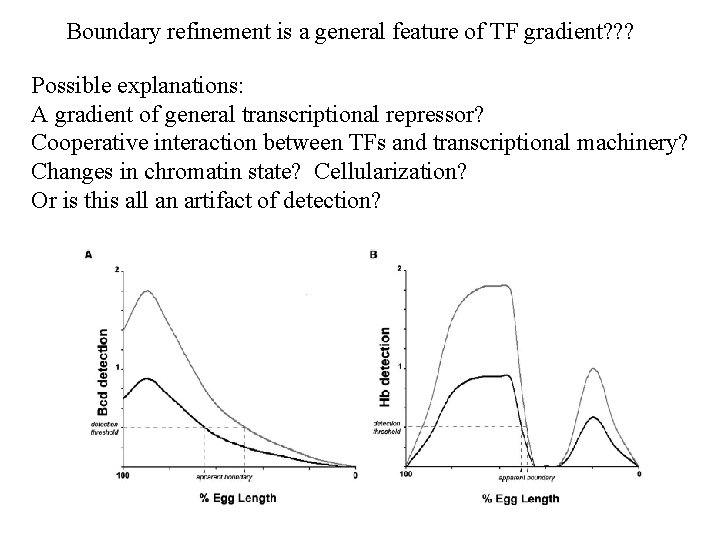
Boundary refinement is a general feature of TF gradient? ? ? Possible explanations: A gradient of general transcriptional repressor? Cooperative interaction between TFs and transcriptional machinery? Changes in chromatin state? Cellularization? Or is this all an artifact of detection?
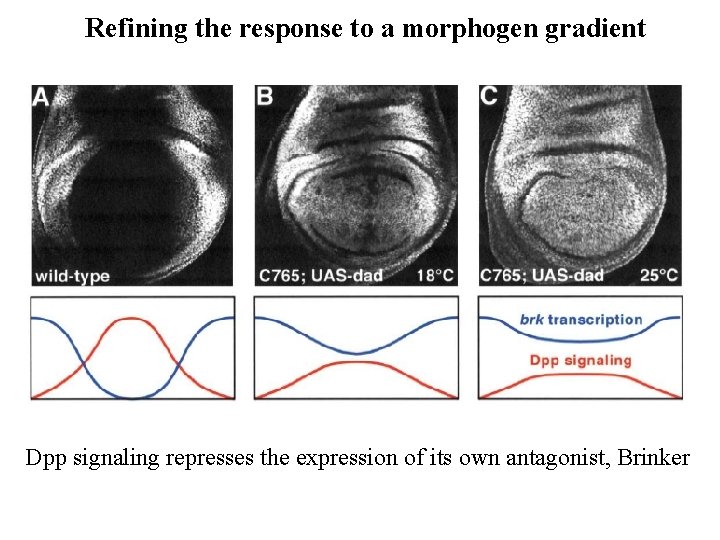
Refining the response to a morphogen gradient Dpp signaling represses the expression of its own antagonist, Brinker
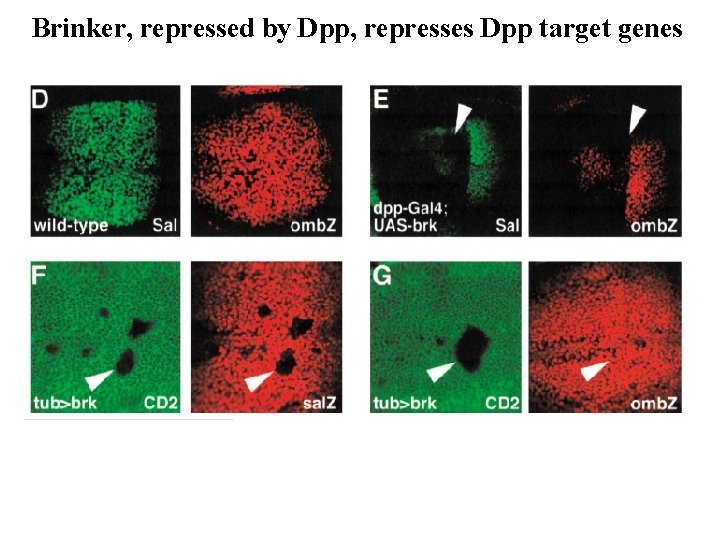
Brinker, repressed by Dpp, represses Dpp target genes
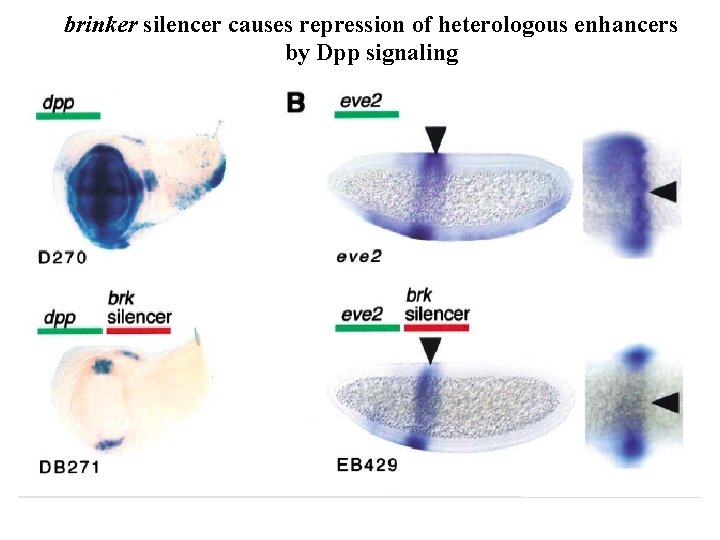
brinker silencer causes repression of heterologous enhancers by Dpp signaling
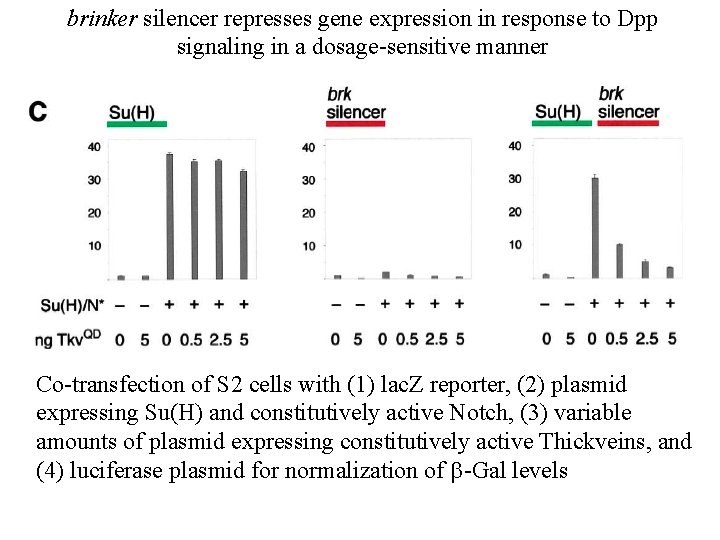
brinker silencer represses gene expression in response to Dpp signaling in a dosage-sensitive manner Co-transfection of S 2 cells with (1) lac. Z reporter, (2) plasmid expressing Su(H) and constitutively active Notch, (3) variable amounts of plasmid expressing constitutively active Thickveins, and (4) luciferase plasmid for normalization of b-Gal levels
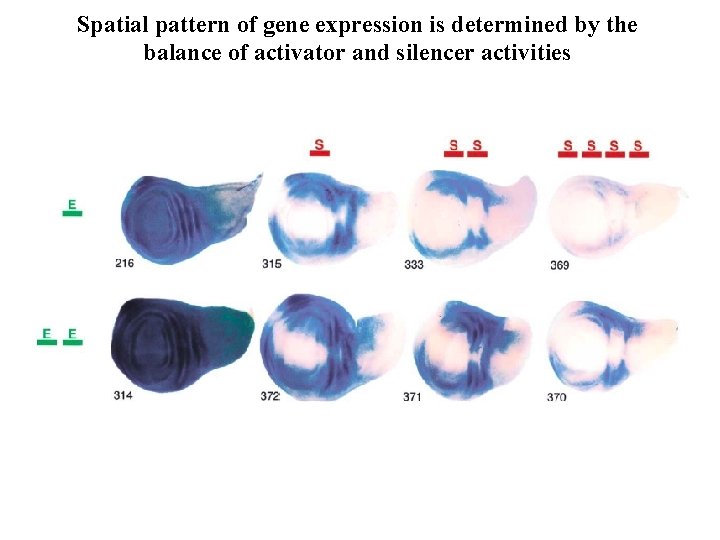
Spatial pattern of gene expression is determined by the balance of activator and silencer activities
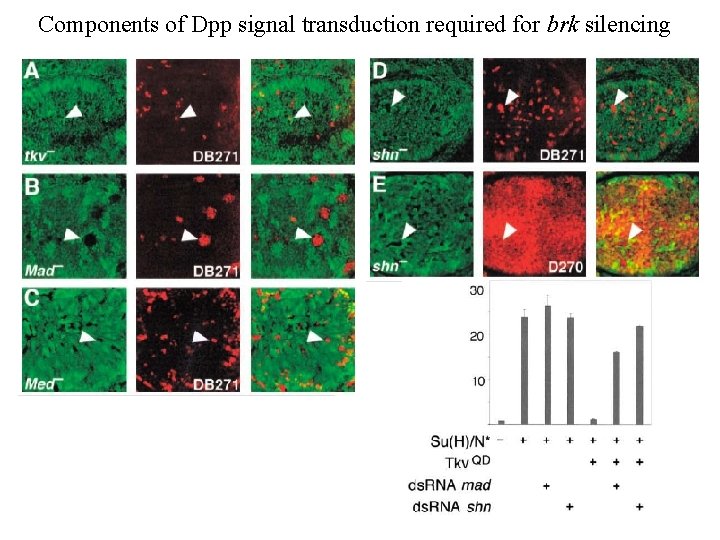
Components of Dpp signal transduction required for brk silencing
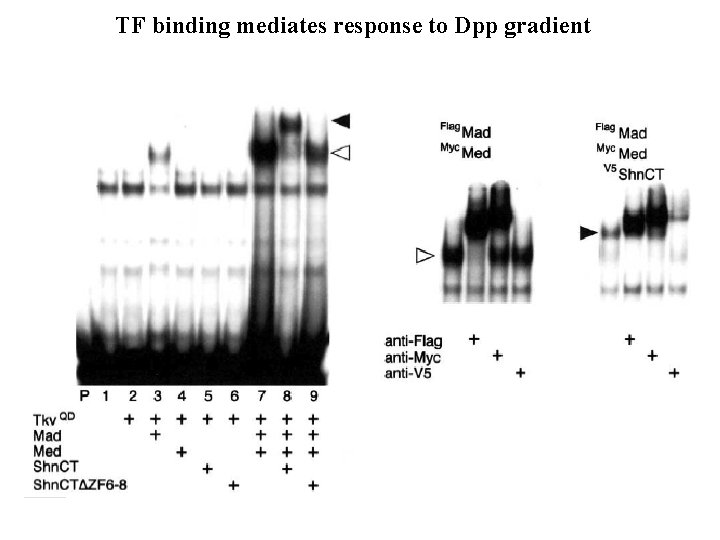
TF binding mediates response to Dpp gradient

This was as far as I got…
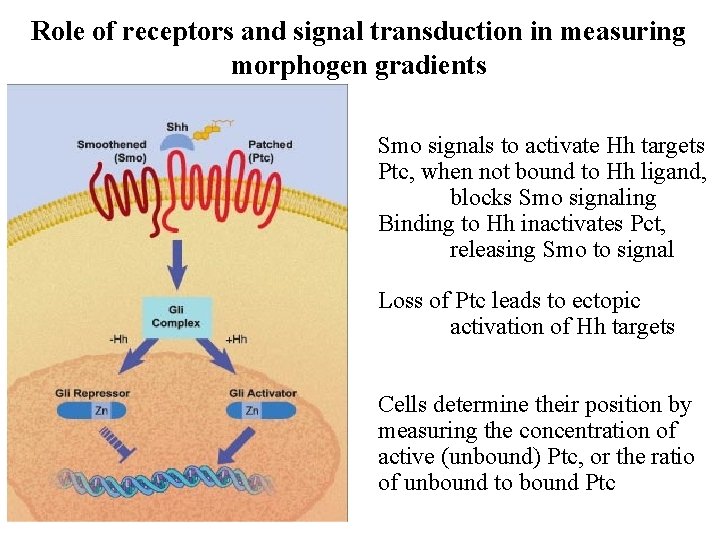
Role of receptors and signal transduction in measuring morphogen gradients Smo signals to activate Hh targets Ptc, when not bound to Hh ligand, blocks Smo signaling Binding to Hh inactivates Pct, releasing Smo to signal Loss of Ptc leads to ectopic activation of Hh targets Cells determine their position by measuring the concentration of active (unbound) Ptc, or the ratio of unbound to bound Ptc
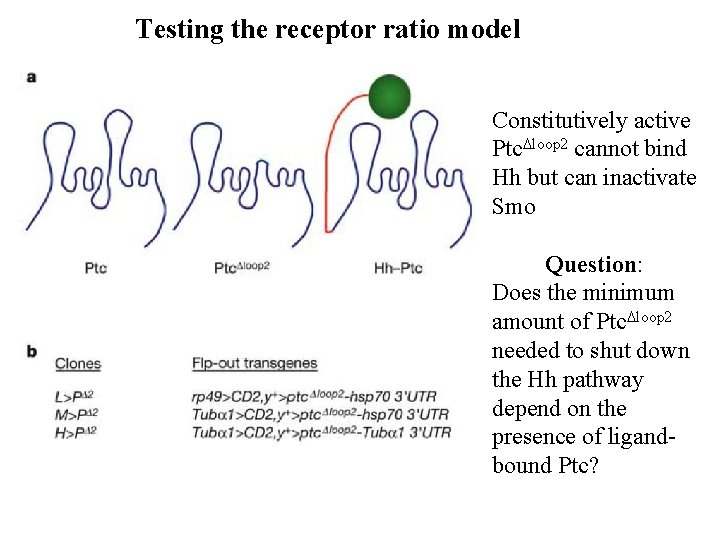
Testing the receptor ratio model Constitutively active Ptc. Dloop 2 cannot bind Hh but can inactivate Smo Question: Does the minimum amount of Ptc. Dloop 2 needed to shut down the Hh pathway depend on the presence of ligandbound Ptc?
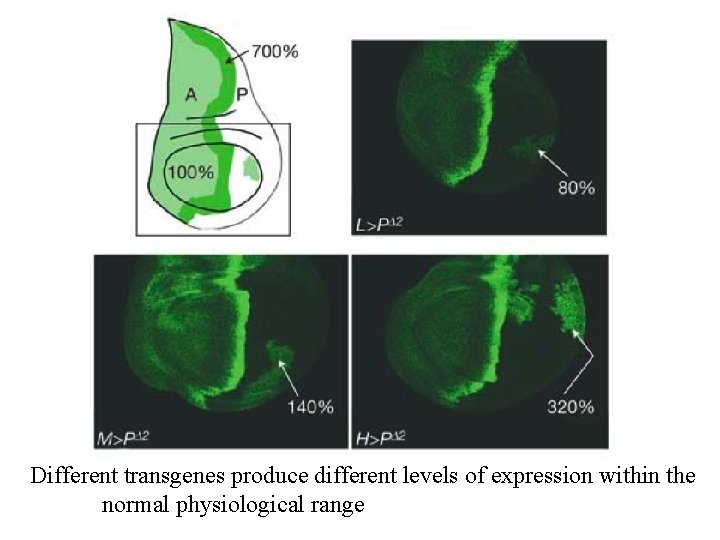
Different transgenes produce different levels of expression within the normal physiological range
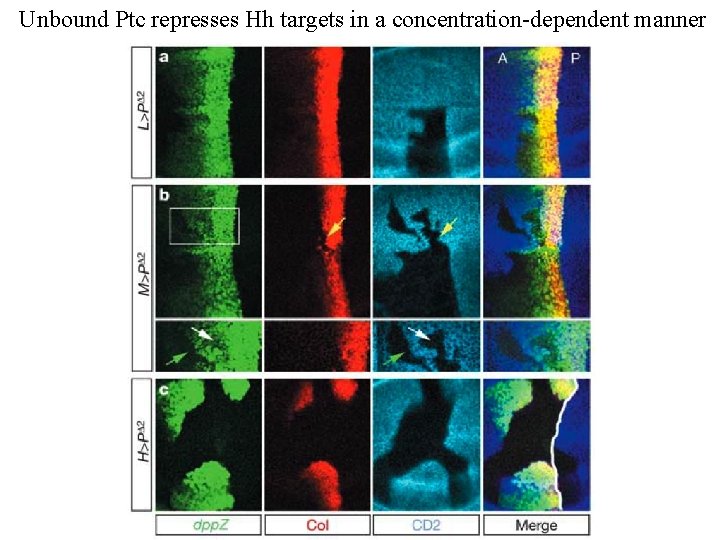
Unbound Ptc represses Hh targets in a concentration-dependent manner
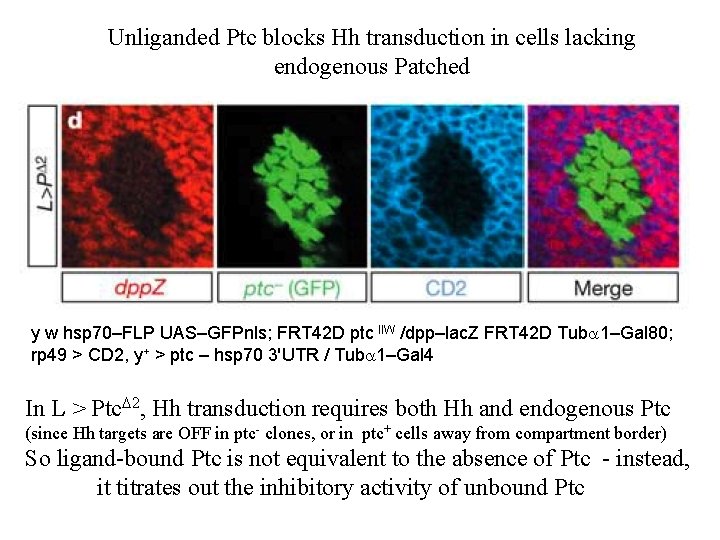
Unliganded Ptc blocks Hh transduction in cells lacking endogenous Patched y w hsp 70–FLP UAS–GFPnls; FRT 42 D ptc IIW /dpp–lac. Z FRT 42 D Tuba 1–Gal 80; rp 49 > CD 2, y+ > ptc – hsp 70 3'UTR / Tuba 1–Gal 4 In L > Ptc. D 2, Hh transduction requires both Hh and endogenous Ptc (since Hh targets are OFF in ptc- clones, or in ptc+ cells away from compartment border) So ligand-bound Ptc is not equivalent to the absence of Ptc - instead, it titrates out the inhibitory activity of unbound Ptc
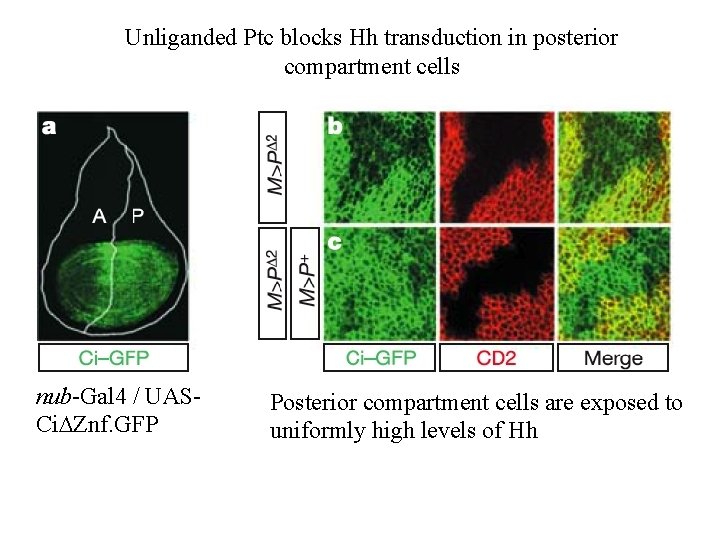
Unliganded Ptc blocks Hh transduction in posterior compartment cells nub-Gal 4 / UASCi. DZnf. GFP Posterior compartment cells are exposed to uniformly high levels of Hh
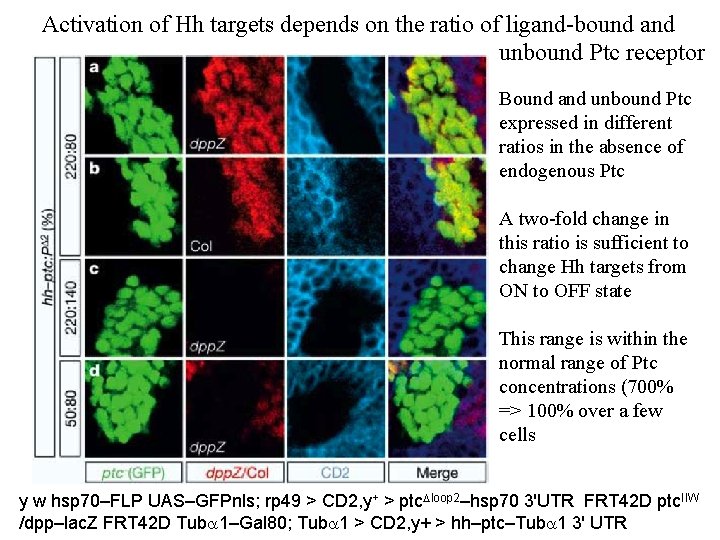
Activation of Hh targets depends on the ratio of ligand-bound and unbound Ptc receptor Bound and unbound Ptc expressed in different ratios in the absence of endogenous Ptc A two-fold change in this ratio is sufficient to change Hh targets from ON to OFF state This range is within the normal range of Ptc concentrations (700% => 100% over a few cells y w hsp 70–FLP UAS–GFPnls; rp 49 > CD 2, y+ > ptc. Dloop 2–hsp 70 3'UTR FRT 42 D ptc. IIW /dpp–lac. Z FRT 42 D Tuba 1–Gal 80; Tuba 1 > CD 2, y+ > hh–ptc–Tuba 1 3' UTR
- Slides: 40