Positional information fields boundaries and gradients Development requires

Positional information: fields, boundaries, and gradients Development requires a dramatic increase in the amount of information contained within the organism. The "new" information is contained in the genome, and is gradually translated into cellular processes. The principal ways in which this happens is by (1) subdivision of larger fields of cells into smaller fields, and (2) specifying the "address" of each cell within the field. This is a recursive process that requires translation of gradients of gene expression into sharp boundaries, and initiation of new gradients by these boundaries
Specification of cell fates Positional cues (Pattern formation): Cell fate is determined by its spatial position within a morphogenetic field during a critical time period Historical cues (Cell lineage): Cell fate is determined by inherited molecules or gene expression states Both mechanisms are required for cell specification, and often act simultaneously.
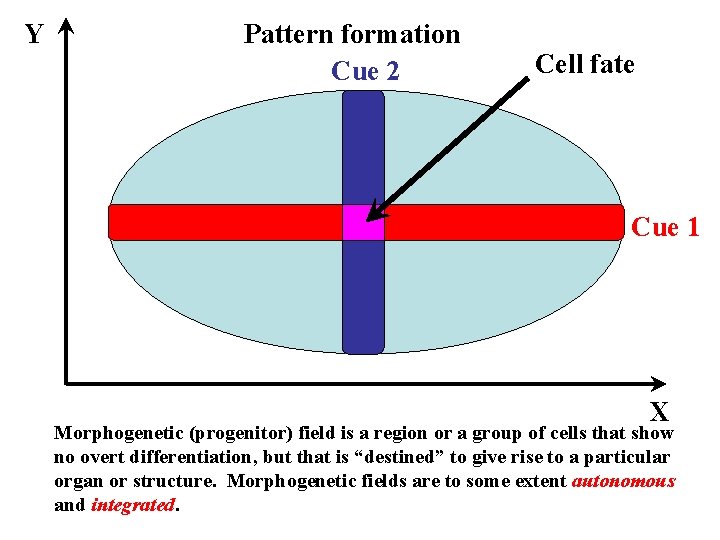
Y Pattern formation Cue 2 Cell fate Cue 1 X Morphogenetic (progenitor) field is a region or a group of cells that show no overt differentiation, but that is “destined” to give rise to a particular organ or structure. Morphogenetic fields are to some extent autonomous and integrated.
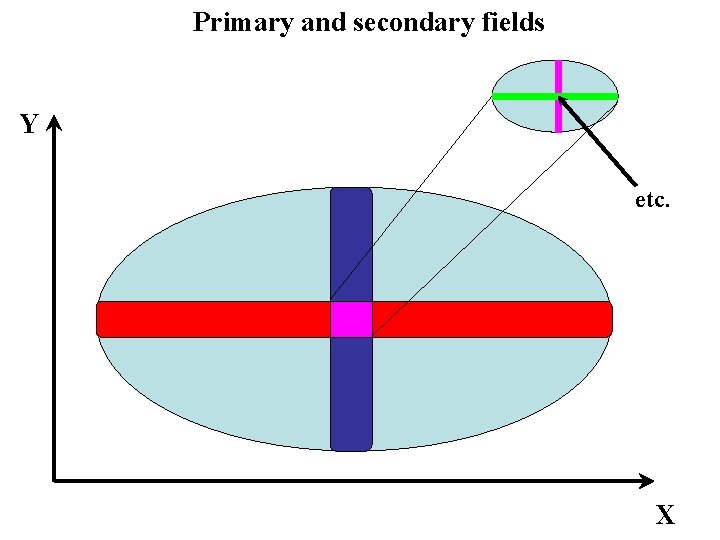
Primary and secondary fields Y etc. X
Morphogen gradients A morphogen is a (usually) secreted molecule that induces cell fate decisions in recipient cells in a concentration-dependent manner Requires: Spatially restricted production Long-range distribution (passive or active) Reception and interpretation Interpretation is context-dependent Most animal morphogens belong to a small number of well-conserved and widely distributed families
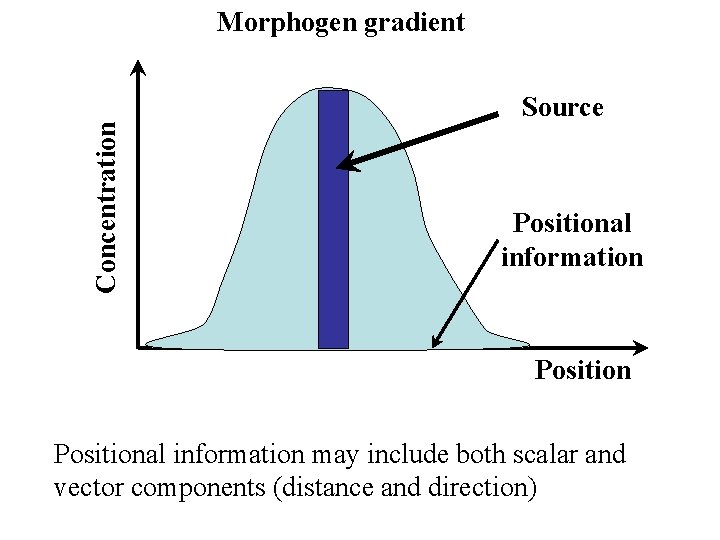
Concentration Morphogen gradient Source Positional information may include both scalar and vector components (distance and direction)
Gradients and cell polarity
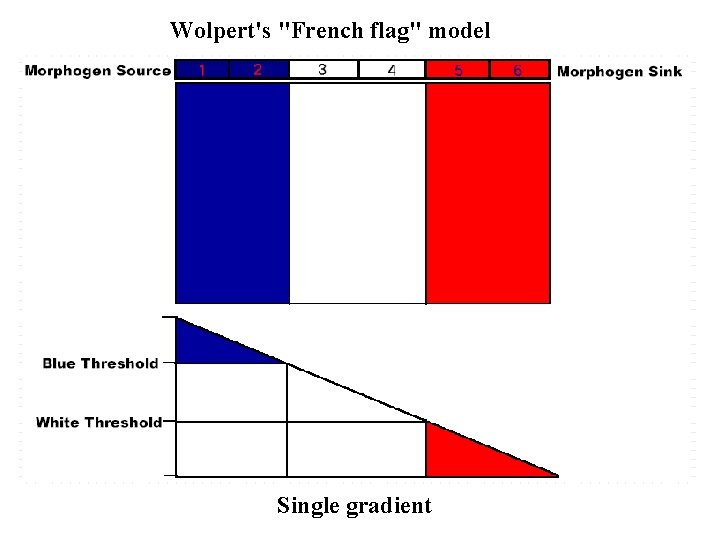
Wolpert's "French flag" model Single gradient
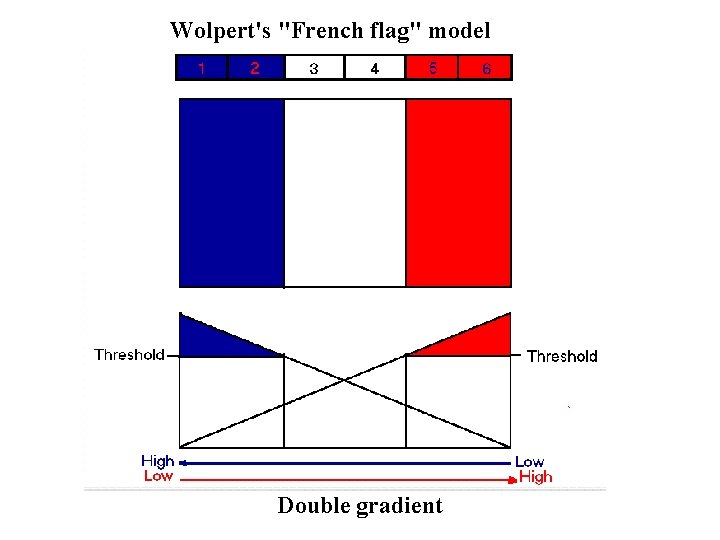
Wolpert's "French flag" model Double gradient
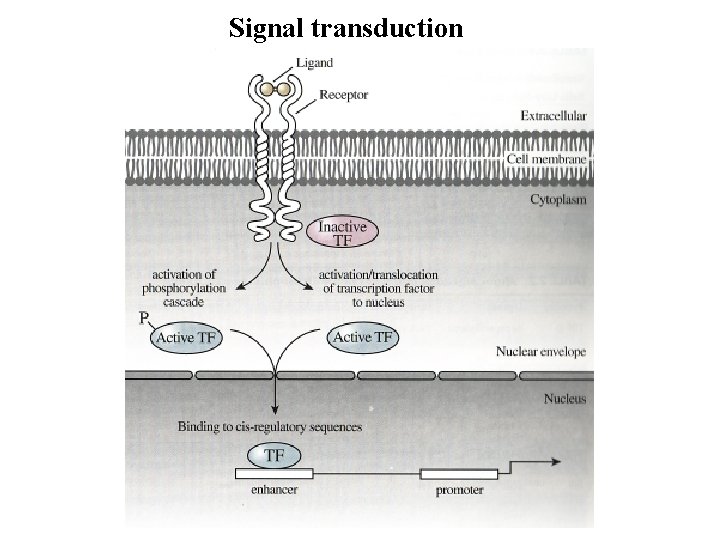
Signal transduction
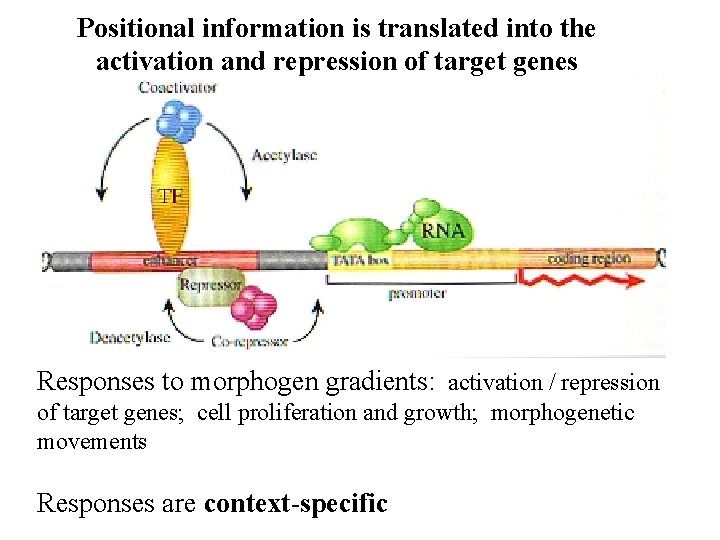
Positional information is translated into the activation and repression of target genes Responses to morphogen gradients: activation / repression of target genes; cell proliferation and growth; morphogenetic movements Responses are context-specific
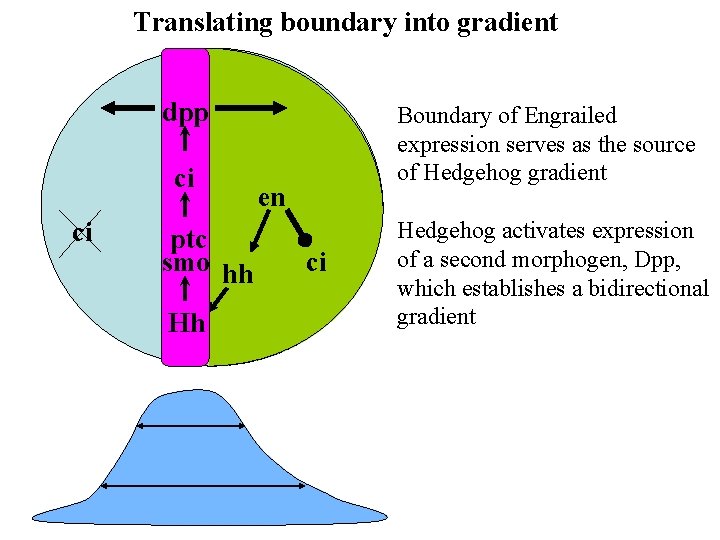
Translating boundary into gradient dpp ci ci ptc smo hh Hh Boundary of Engrailed expression serves as the source of Hedgehog gradient en ci Hedgehog activates expression of a second morphogen, Dpp, which establishes a bidirectional gradient
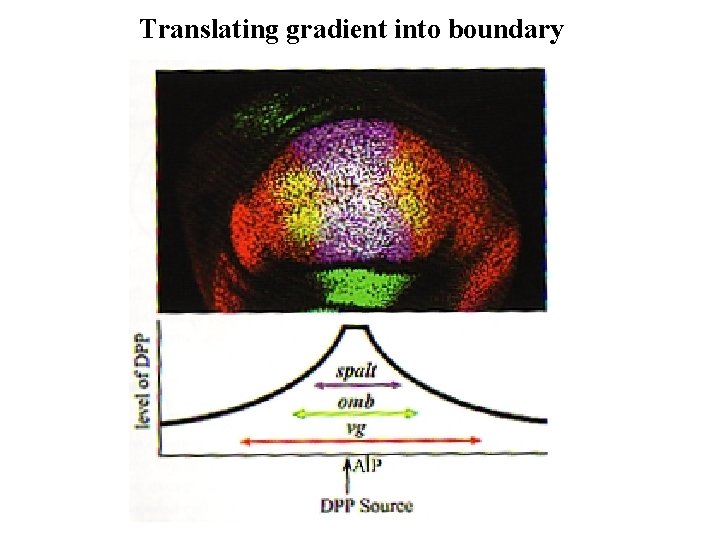
Translating gradient into boundary
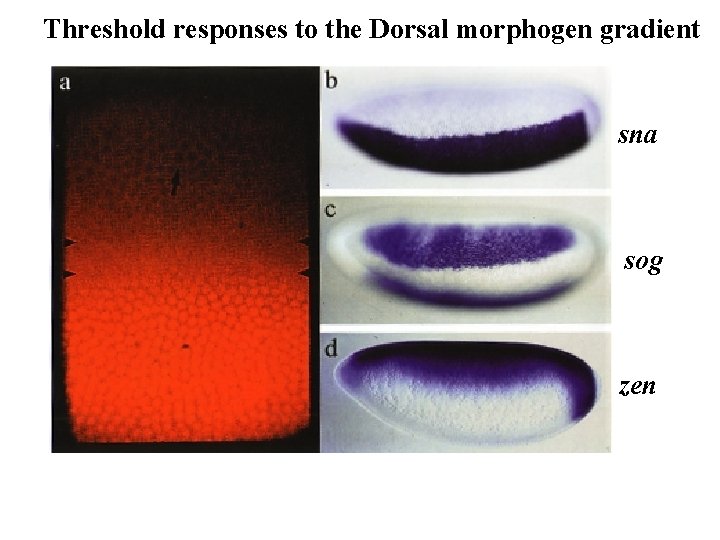
Threshold responses to the Dorsal morphogen gradient sna sog zen
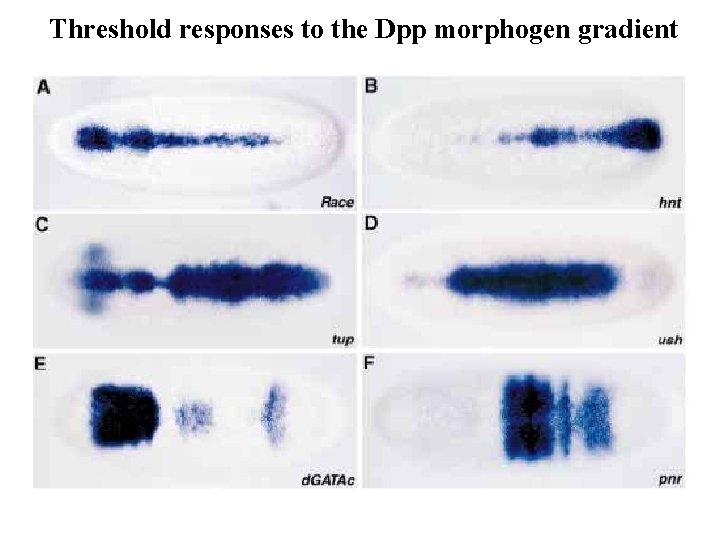
Threshold responses to the Dpp morphogen gradient
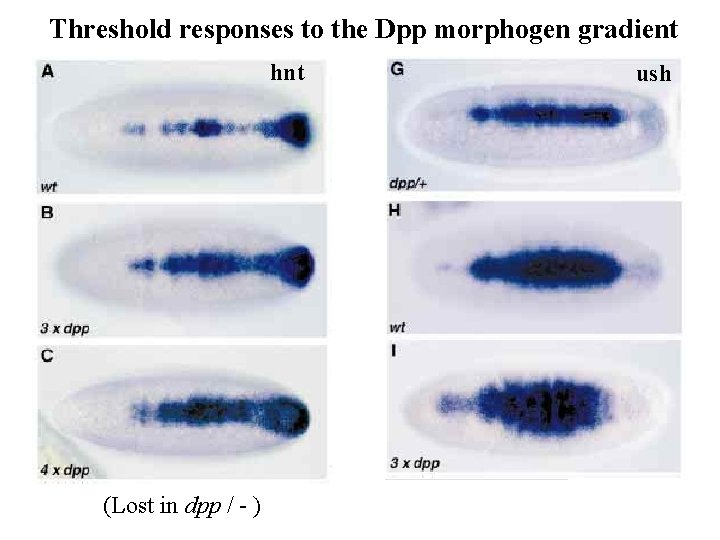
Threshold responses to the Dpp morphogen gradient hnt (Lost in dpp / - ) ush
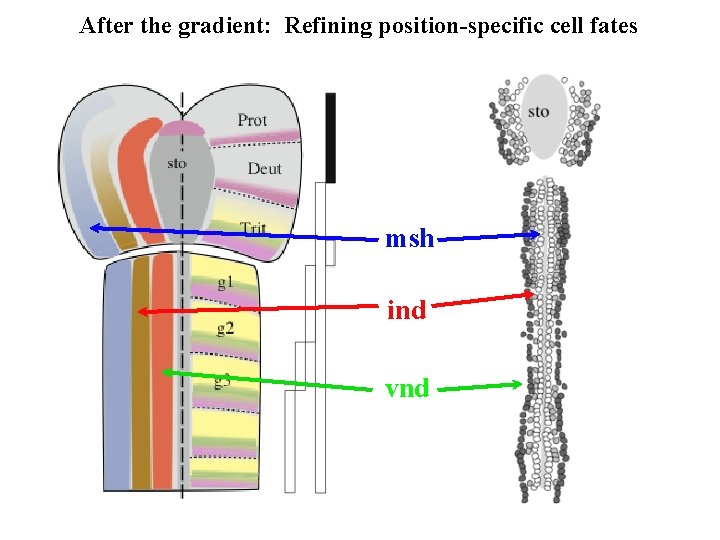
After the gradient: Refining position-specific cell fates msh ind vnd
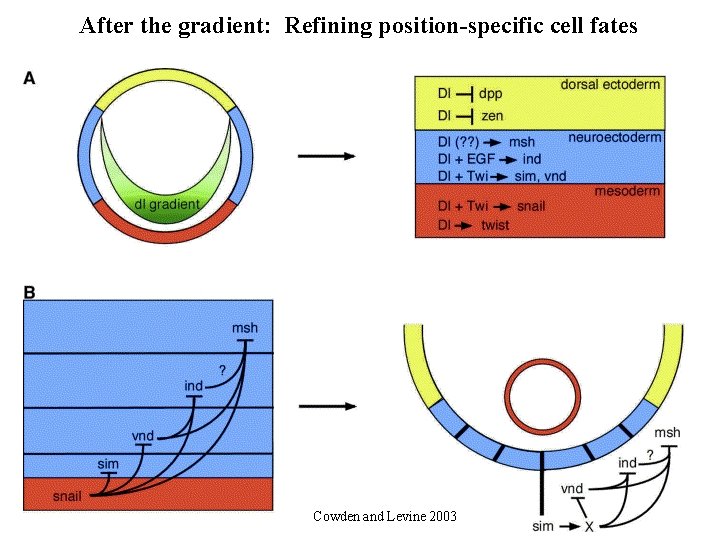
After the gradient: Refining position-specific cell fates Cowden and Levine 2003
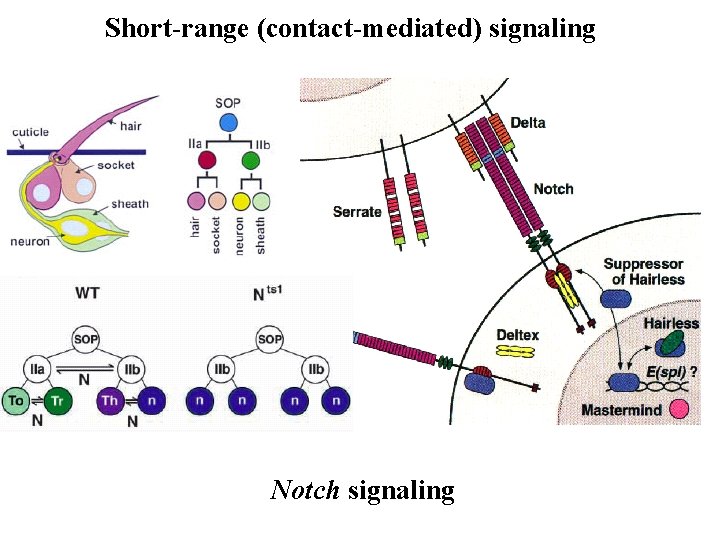
Short-range (contact-mediated) signaling Notch signaling
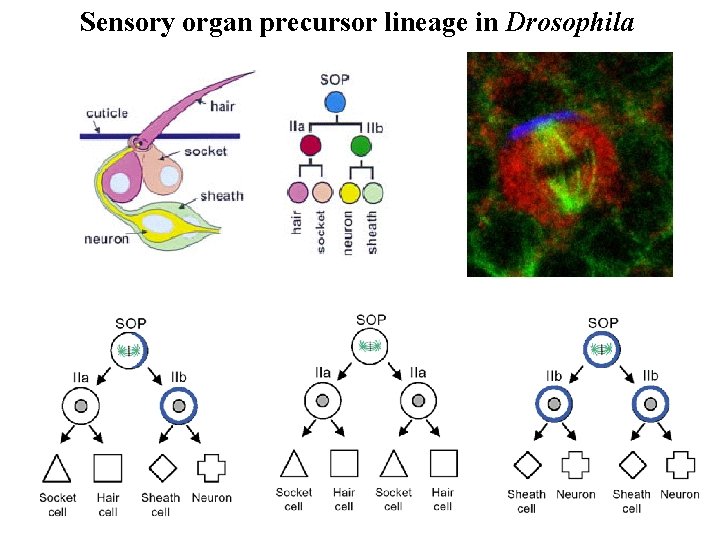
Sensory organ precursor lineage in Drosophila
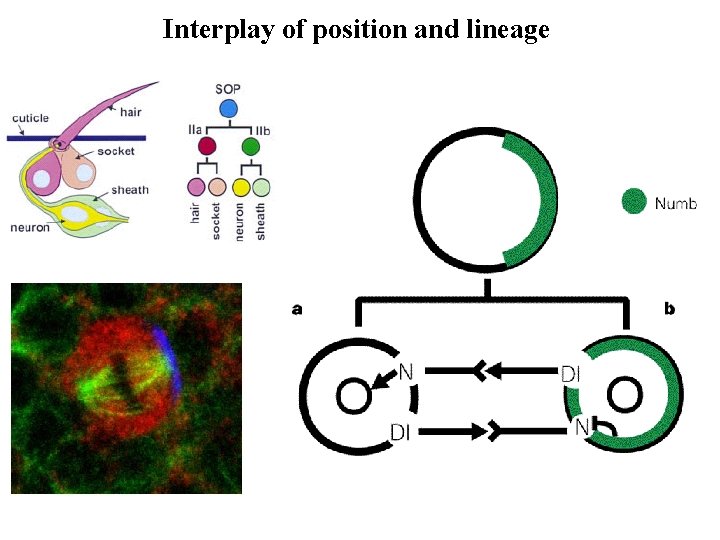
Interplay of position and lineage
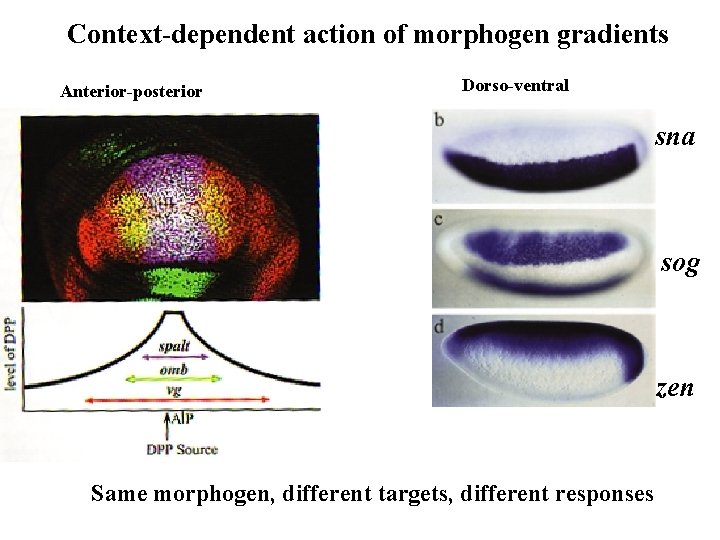
Context-dependent action of morphogen gradients Anterior-posterior Dorso-ventral sna sog zen Same morphogen, different targets, different responses
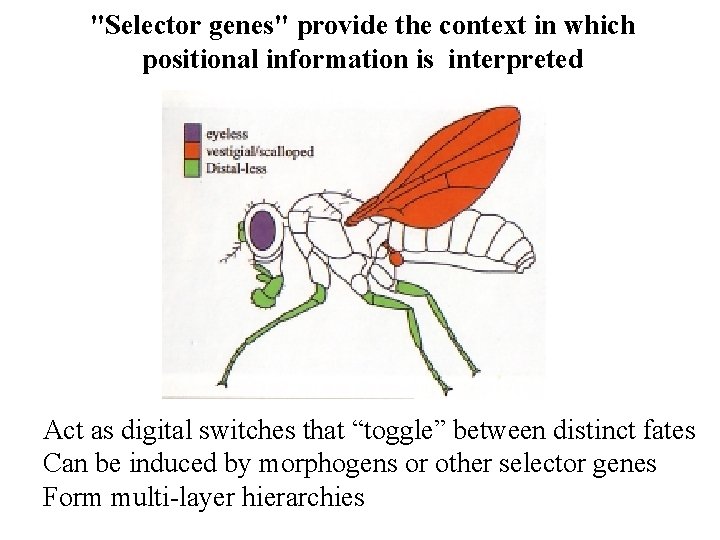
"Selector genes" provide the context in which positional information is interpreted Act as digital switches that “toggle” between distinct fates Can be induced by morphogens or other selector genes Form multi-layer hierarchies
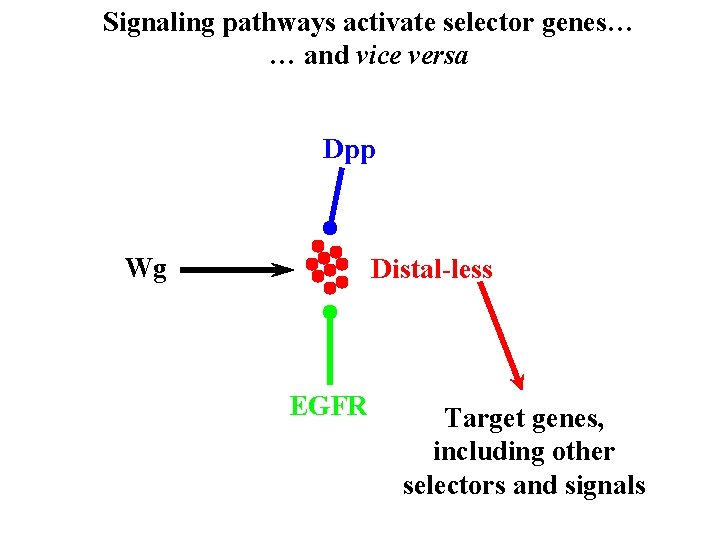
Signaling pathways activate selector genes… … and vice versa Dpp Wg Distal-less EGFR Target genes, including other selectors and signals
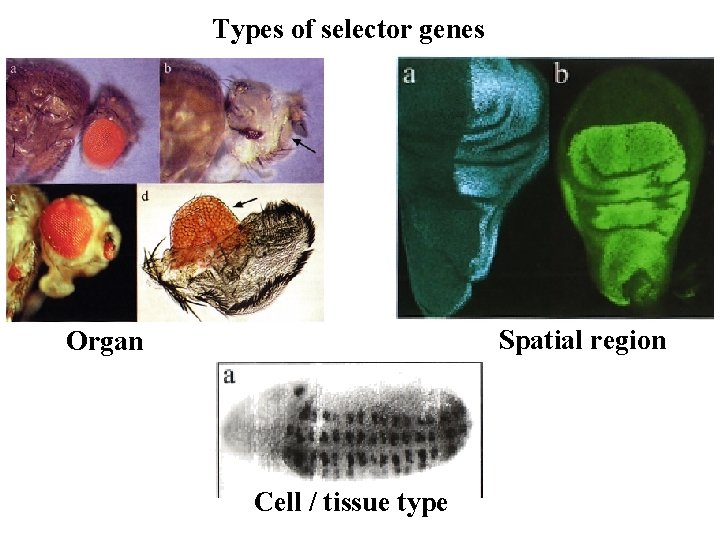
Types of selector genes Spatial region Organ Cell / tissue type
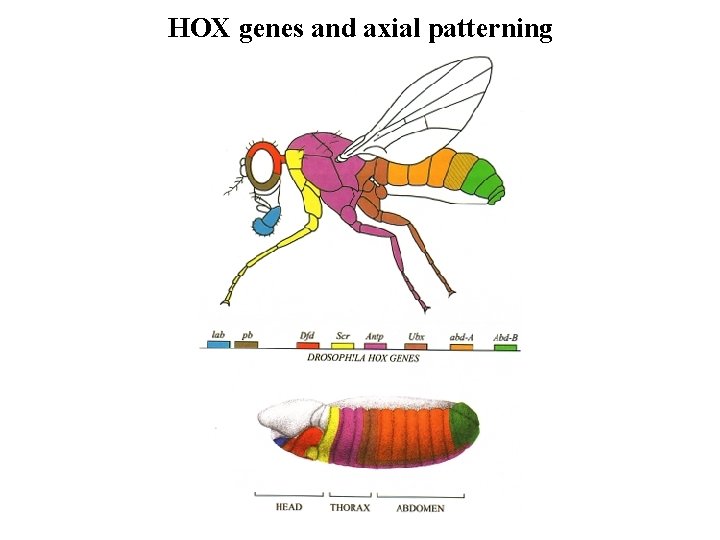
HOX genes and axial patterning
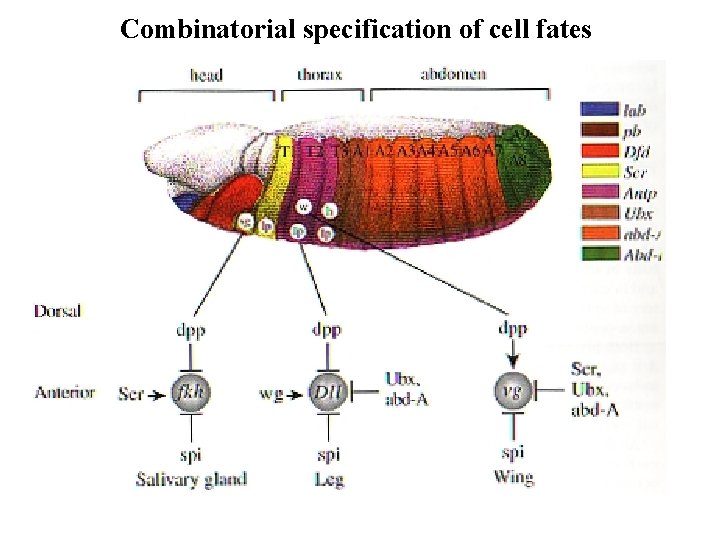
Combinatorial specification of cell fates
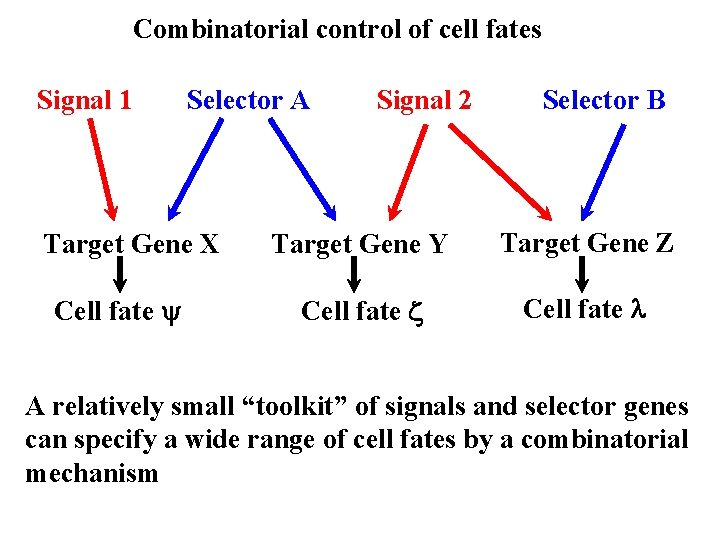
Combinatorial control of cell fates Signal 1 Selector A Target Gene X Cell fate y Signal 2 Selector B Target Gene Y Target Gene Z Cell fate z Cell fate l A relatively small “toolkit” of signals and selector genes can specify a wide range of cell fates by a combinatorial mechanism
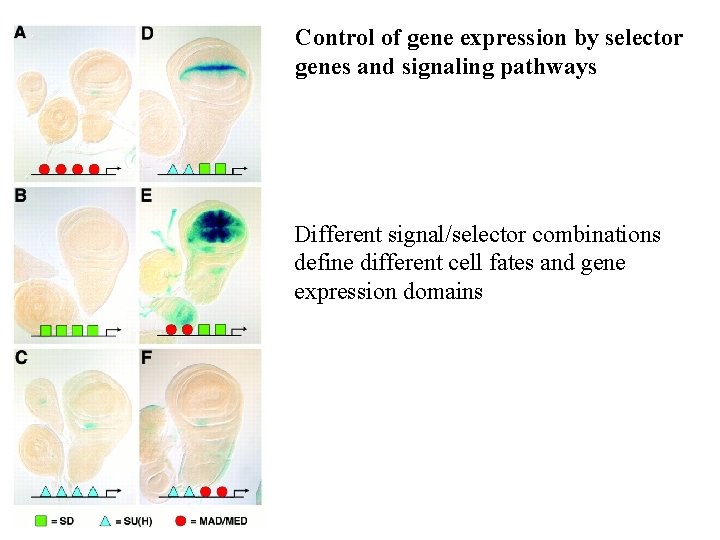
Control of gene expression by selector genes and signaling pathways Different signal/selector combinations define different cell fates and gene expression domains
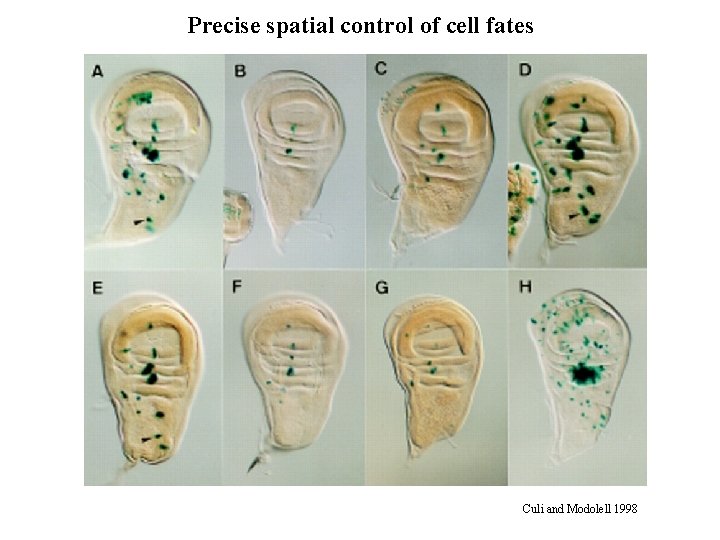
Precise spatial control of cell fates Culi and Modolell 1998
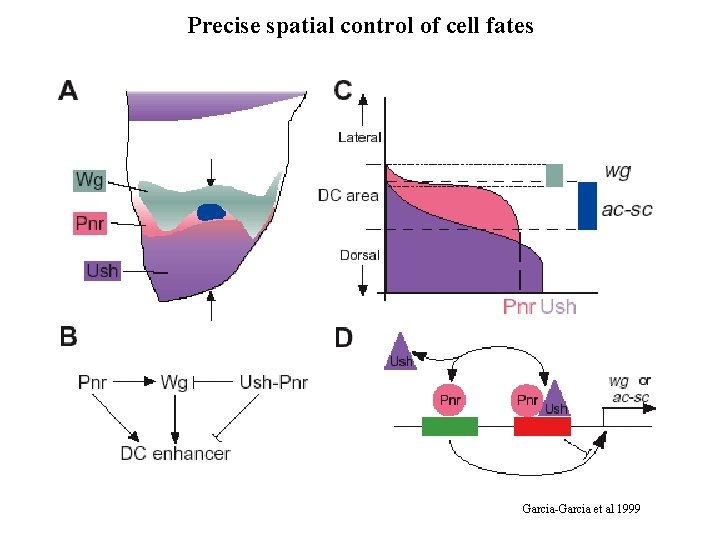
Precise spatial control of cell fates Garcia-Garcia et al 1999
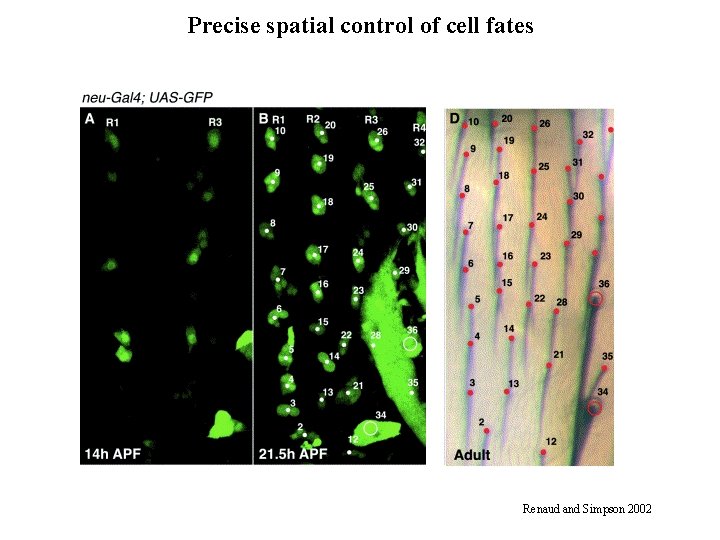
Precise spatial control of cell fates Renaud and Simpson 2002
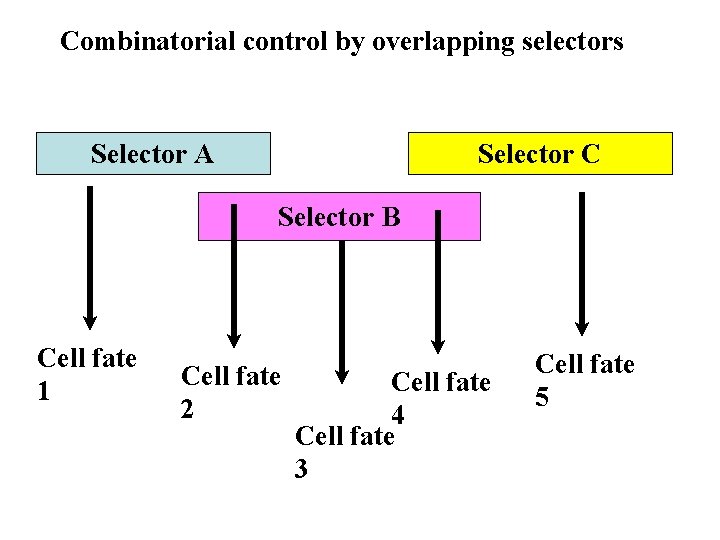
Combinatorial control by overlapping selectors Selector A Selector C Selector B Cell fate 1 Cell fate 2 Cell fate 4 Cell fate 3 Cell fate 5
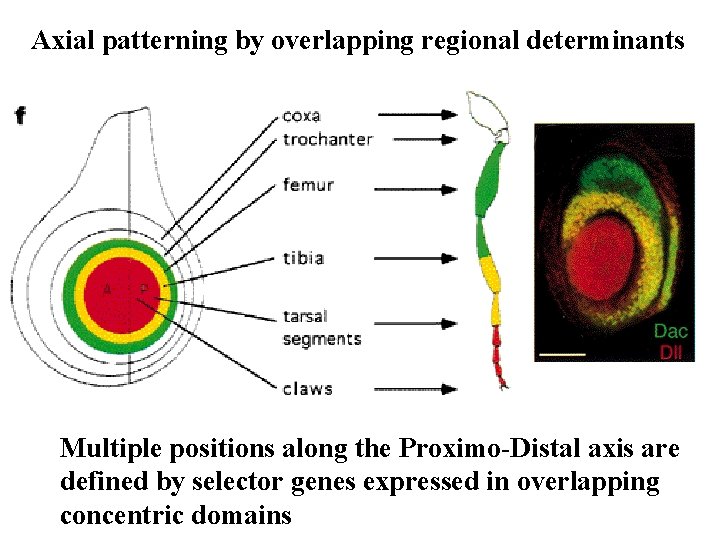
Axial patterning by overlapping regional determinants Multiple positions along the Proximo-Distal axis are defined by selector genes expressed in overlapping concentric domains
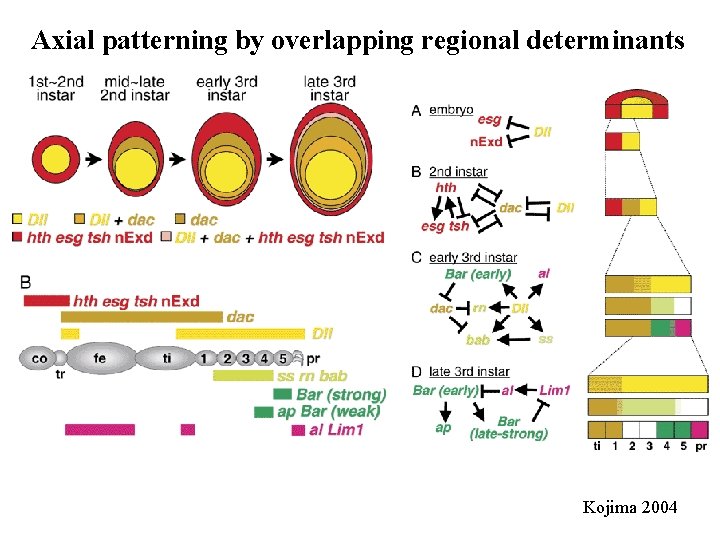
Axial patterning by overlapping regional determinants Kojima 2004
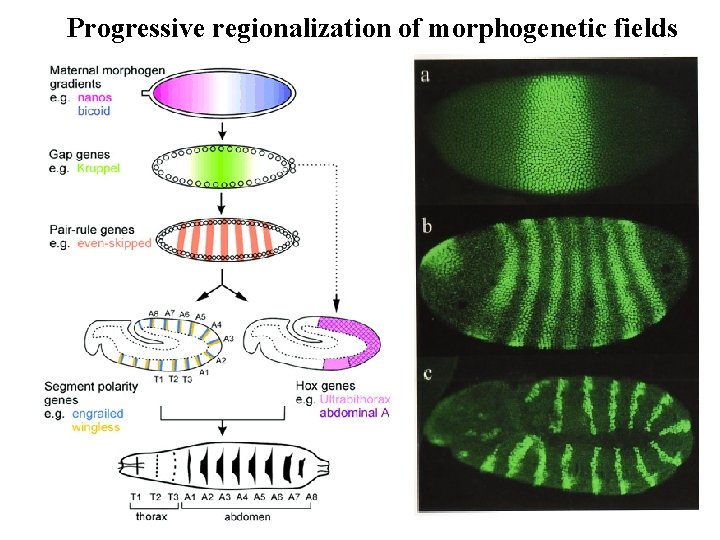
Progressive regionalization of morphogenetic fields
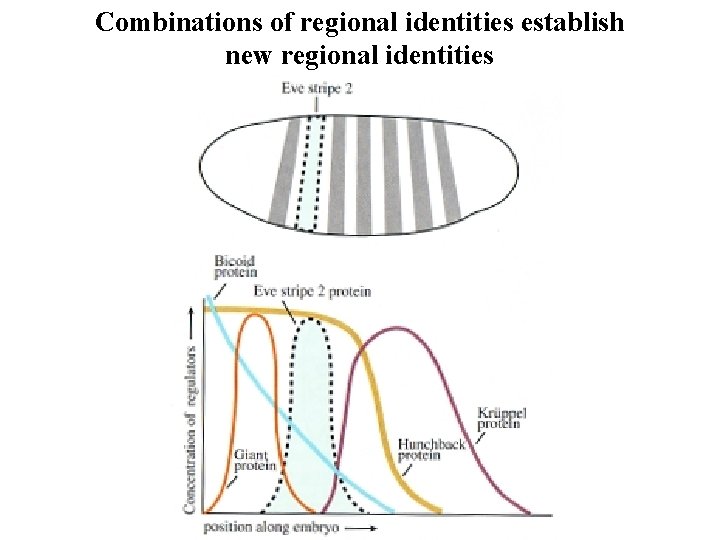
Combinations of regional identities establish new regional identities
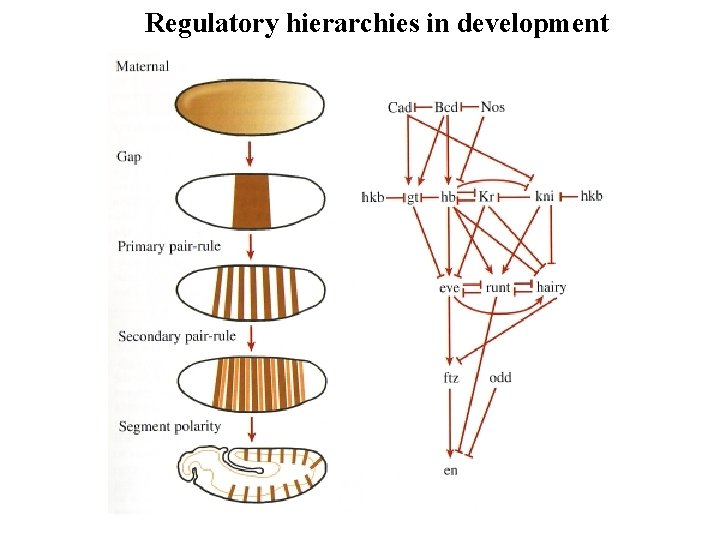
Regulatory hierarchies in development
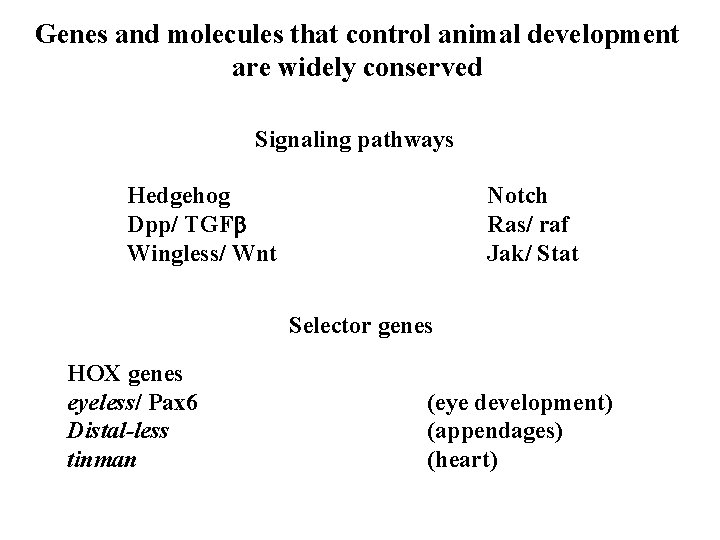
Genes and molecules that control animal development are widely conserved Signaling pathways Hedgehog Dpp/ TGFb Wingless/ Wnt Notch Ras/ raf Jak/ Stat Selector genes HOX genes eyeless/ Pax 6 Distal-less tinman (eye development) (appendages) (heart)
- Slides: 39