Polyphasic taxonomy of marine bacteria from the SAR
Polyphasic taxonomy of marine bacteria from the SAR 11 group Ia: Pelagibacter ubiquis (strain HTCC 1062) & Pelagibacter bermudensis (strain HTCC 7211) Sarah N. Brown Dr. Stephen Giovannoni Dr. Jang-Cheon Cho Department of Microbiology HHMI 2011 3 -D structures of Pelagibacter ubique (2006) (Pelagibacter means "bacterium of the sea")
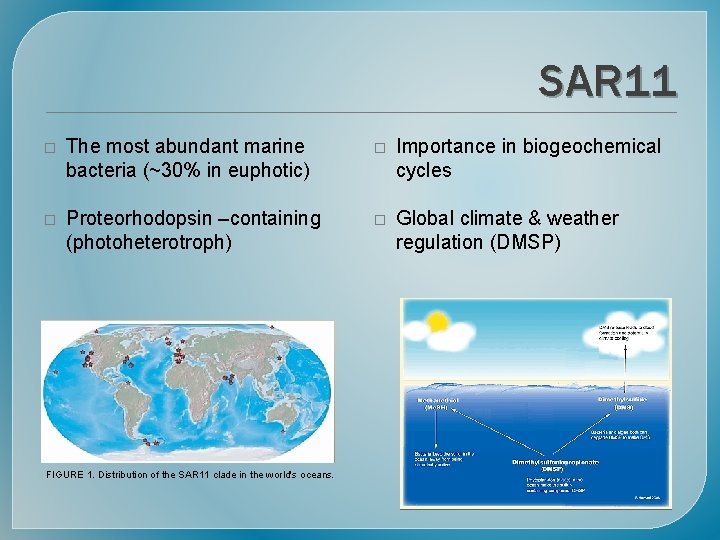
SAR 11 � The most abundant marine bacteria (~30% in euphotic) � Importance in biogeochemical cycles � Proteorhodopsin –containing (photoheterotroph) � Global climate & weather regulation (DMSP) FIGURE 1. Distribution of the SAR 11 clade in the world's oceans.

Background � In 2002, SAR 11 cells were first isolated in seawater-based medium (no colonies produced). � Growth of Pelagibacter in artificial seawater medium (ASW) is a recent advancement. � Non-colony forming property and oligotrophy made it difficult for taxonomy � Purpose: To characterize & provide official nomenclature for SAR 11 (strains 1062 & 7211) • SAR 11: Candidatus Pelagibacter Fig. 4. Cultures of oligotrophic marine bacteria growing in carboys of autoclaved seawater.

Polyphasic Taxonomy � Incorporates multiple methods for identification & description of new species � Species: the basic unit of bacterial taxonomy � SAR 11 clade • Class: Alphaproteobacteria GENOTYPIC INFORMATION PHENOTYPIC INFORMATION
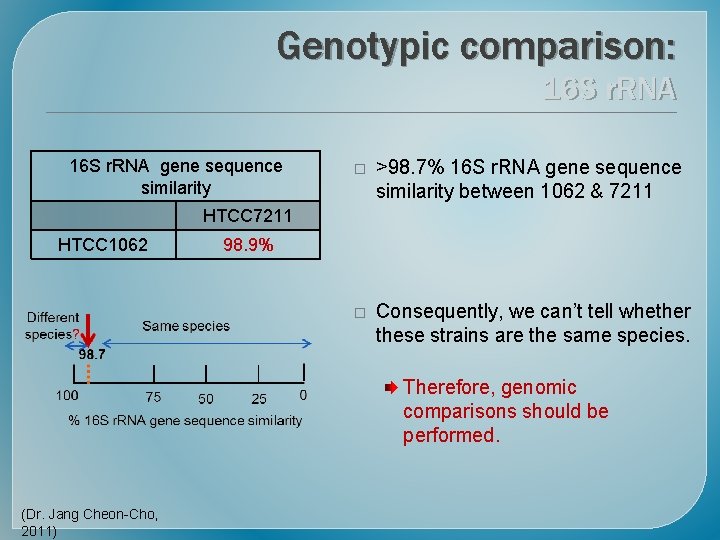
Genotypic comparison: 16 S r. RNA gene sequence similarity � >98. 7% 16 S r. RNA gene sequence similarity between 1062 & 7211 � Consequently, we can’t tell whether these strains are the same species. HTCC 7211 HTCC 1062 98. 9% Therefore, genomic comparisons should be performed. (Dr. Jang Cheon-Cho, 2011)

ANI: A method for bacterial species demarcation � >95 -96% average nucleotide identity (ANIb/ANIm) indicates ‘true (same) species’ Different species <95 -96% ANIm/b Same species Fig. , m= MUMmer computer algorithm; b= BLAST algorithm (Dr. Jang Cheon-Cho, 2011) ANIb HTCC 1062 HTCC 7211 76. 73 ANIm HTCC 1062 HTCC 7211 82. 61 Strains show <95 -96% average nucleotide identity (ANIb/m) Therefore, HTCC 1062 & HTCC 7211 represent separate genomic species

Conclusion of genotypic analysis: 2 species HTCC 1062 P. ubiquis HTCC 1002 HTCC 7211 P. bermudensis HIMB 5 0. 001 1. HTCC 1062→ Pelagibacter ubiquis gen. nov. , sp. nov. 2. HTCC 7211→ Pelagibacter bermudensis sp. nov. (Dr. Jang Cheon-Cho, 2011) Group Ia

Phenotypic comparisons: growth conditions � Prepare artificial seawater medium (ASW) • Salinity: w/out Na. Cl & w/10% Na. Cl • p. H: adjust w/0. 1 M Na. OH & 0. 1 M HCl � Add nutrients & inoculum � Dispense into 156 flasks (triplicates of each growth condition) � Incubate • Temp (°C): 4, 8, 12, 16, 20, 23, 25, & 30 • p. H & salinity: 16ºC � Screen for growth
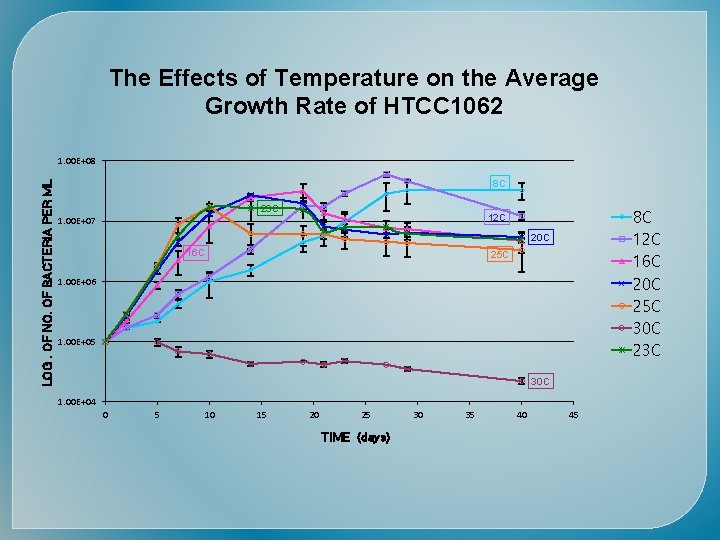
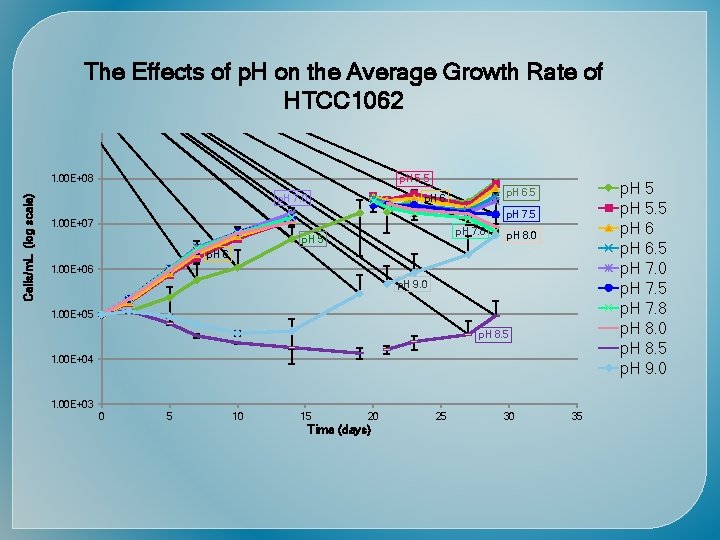
The Effects of Temperature on the Average Growth Rate of HTCC 1062 LOG. OF NO. OF BACTERIA PER ML 1. 00 E+08 8 C 23 C 8 C 12 C 16 C 20 C 25 C 30 C 23 C 12 C 1. 00 E+07 20 C 16 C 25 C 1. 00 E+06 1. 00 E+05 30 C 1. 00 E+04 0 5 10 15 20 25 TIME (days) 30 35 40 45
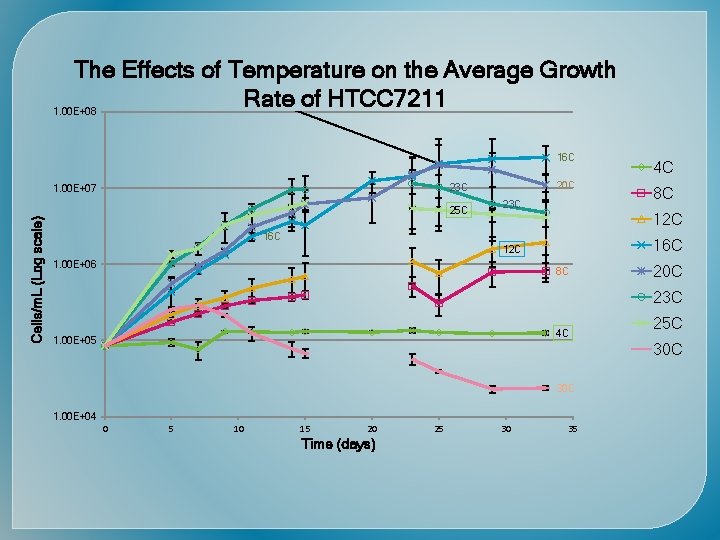
Time (days) Cells/m. L (Log scale)
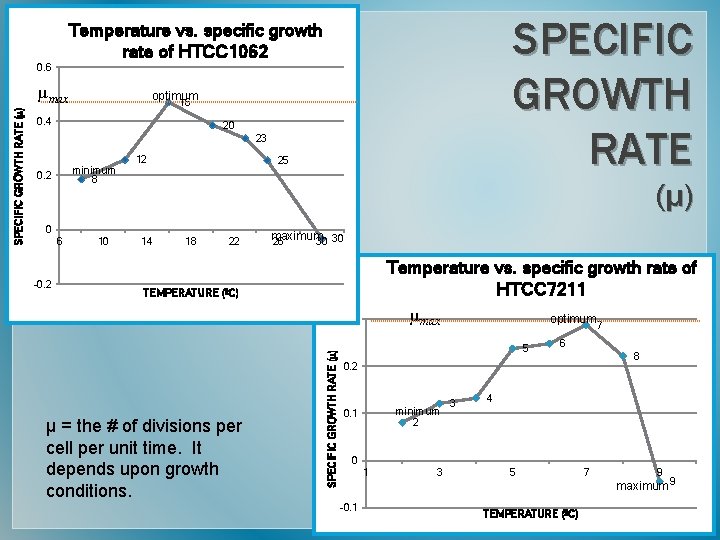
0. 6 µmax optimum 16 0. 4 20 23 minimum 0. 2 12 25 8 (µ) 0 6 -0. 2 10 14 18 22 maximum 26 30 30 Temperature vs. specific growth rate of HTCC 7211 µmax optimum 7 TEMPERATURE (ºC) 0. 3 µ = the # of divisions per cell per unit time. It depends upon growth conditions. SPECIFIC GROWTH RATE (µ) SPECIFIC GROWTH RATE Temperature vs. specific growth rate of HTCC 1062 5 6 8 0. 2 minimum 0. 1 3 4 2 0 1 -0. 1 3 5 TEMPERATURE (ºC) 7 9 maximum 9
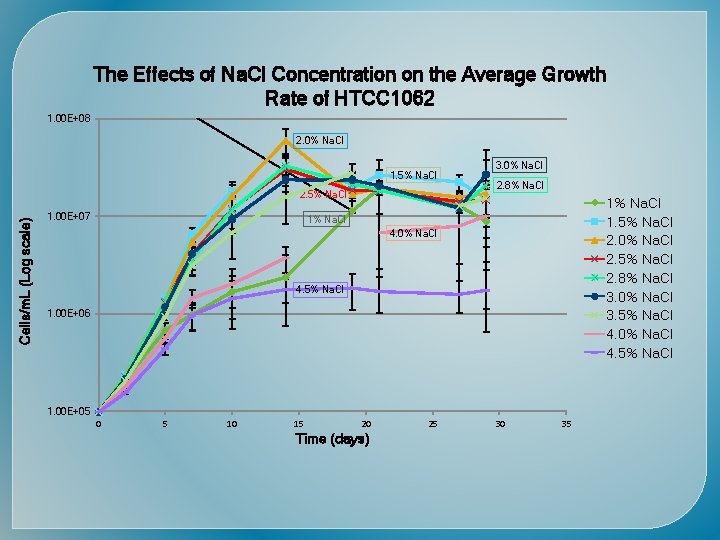
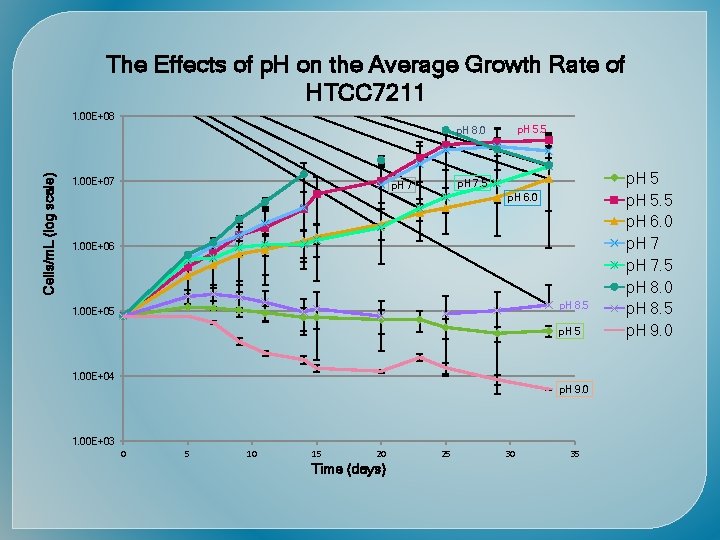
Time (days) Cells/m. L (Log scale)
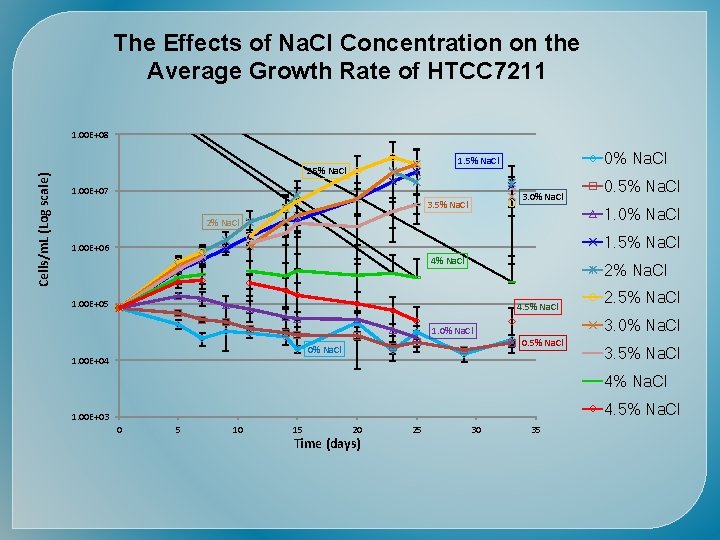
Time (days) Cells/m. L (Log scale)
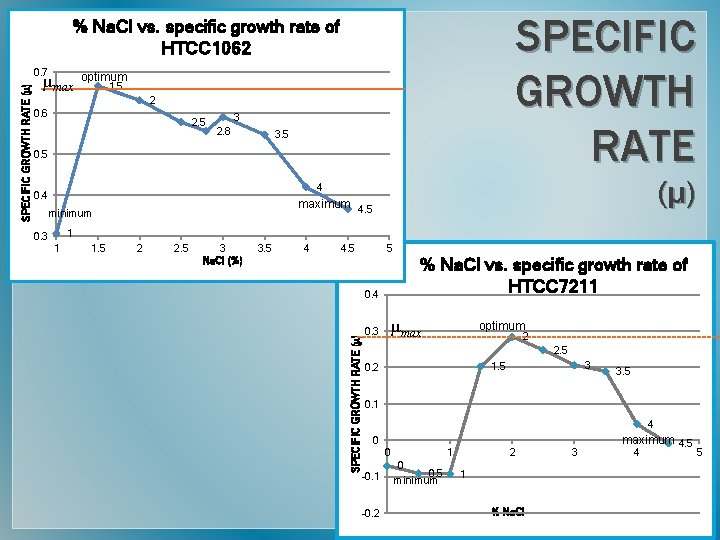
SPECIFIC GROWTH RATE % Na. Cl vs. specific growth rate of HTCC 1062 µmax optimum 1. 5 2 0. 6 2. 5 3 2. 8 3. 5 0. 5 (µ) 4 0. 4 maximum 4. 5 minimum 1 0. 3 1 1. 5 2 2. 5 3 Na. Cl (%) 3. 5 4 4. 5 5 % Na. Cl vs. specific growth rate of HTCC 7211 0. 4 SPECIFIC GROWTH RATE (µ) 0. 7 µmax optimum 0. 3 2 2. 5 3 1. 5 0. 2 3. 5 0. 1 4 0 0 1 0 0. 5 -0. 1 minimum -0. 2 2 1 % Na. Cl 3 maximum 4. 5 4 5
Time (days) Cells/m. L (log scale)
Time (days) Cells/m. L (log scale)
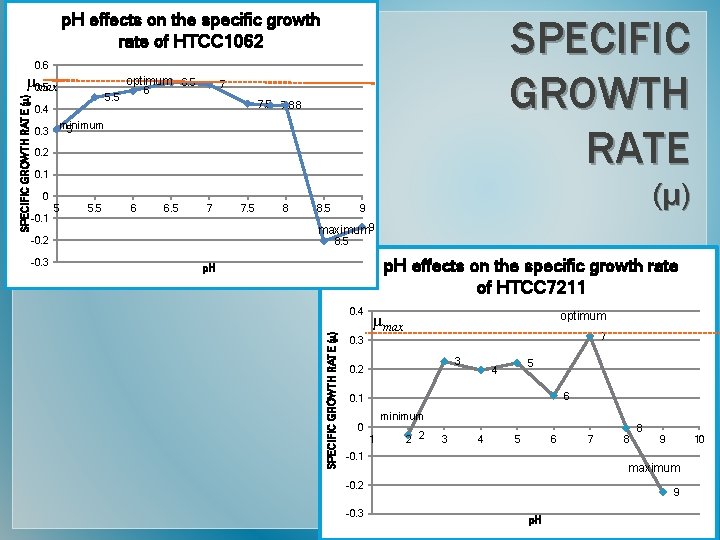
p. H effects on the specific growth rate of HTCC 1062 SPECIFIC GROWTH RATE 0. 6 optimum 6. 5 7 6 5. 5 7. 8 8 0. 4 minimum 5 0. 3 0. 2 0. 1 (µ) 0 -0. 1 5 5. 5 6 6. 5 7 8 8. 5 9 maximum 9 -0. 2 -0. 3 7. 5 8. 5 p. H effects on the specific growth rate of HTCC 7211 p. H 0. 4 SPECIFIC GROWTH RATE (µ) µ 0. 5 max optimum µmax 7 0. 3 3 0. 2 5 4 6 0. 1 minimum 0 1 2 2 3 4 5 6 -0. 1 8 8 9 10 maximum -0. 2 -0. 3 7 9 p. H
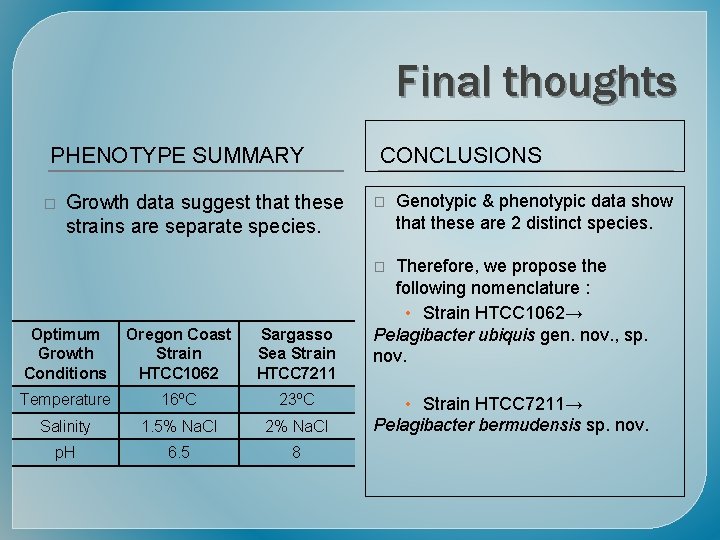
Final thoughts PHENOTYPE SUMMARY � Growth data suggest that these strains are separate species. CONCLUSIONS � Genotypic & phenotypic data show that these are 2 distinct species. Therefore, we propose the following nomenclature : • Strain HTCC 1062→ Pelagibacter ubiquis gen. nov. , sp. nov. � Optimum Growth Conditions Oregon Coast Strain HTCC 1062 Sargasso Sea Strain HTCC 7211 Temperature 16ºC 23ºC Salinity 1. 5% Na. Cl 2% Na. Cl p. H 6. 5 8 • Strain HTCC 7211→ Pelagibacter bermudensis sp. nov.

Acknowledgments � The Gordon & Betty Moore Foundation � Dr. Stephen Giovannoni � Dr. Jang Cheon-Cho � Paul Carini � Kevin Vergin � Giovannoni Lab � HHMI & Dr. Kevin Ahern

References � Konstantinidis, K. , and J. M. Tiedje. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 102: 2567 -2572. � Konstantinidis, K. , and J. M. Tiedje. 2005. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187: 6258 -6264. � Morris, R. M. , Rappé, M. S. , Connon, S. A. , Vergin, K. L. , Siebold, W. A. , Carlson, C. A. , and Giovannoni, S. J. (December 2002). SAR 11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806 -810. doi: 10. 1038/nature 01240. � Nicastro, D. , Schwartz, C. , Pierson, J. , Cho, J. -C. C. , Giovannoni, S. J. , and Mc. Intosh, J. R. (2006). Three-dimensional structure of the tiny bacterium Pelagibacter ubique studied by cryo-electron tomography. Microsc. Microanal. 12(sup 2): 180 -181. � Richter, M. , Rosselló-Móra, R. (October 2009). Shifting the genomic gold standard for the prokaryotic species definition. Biological Sciences - Microbiology: PNAS 106 (45): 19126 -19131; doi: 10. 1073/pnas. 0906412106.
- Slides: 20