Polymerase Chain Reaction PCR v DNA Primer set








- Slides: 23



Polymerase Chain Reaction (PCR) v 중합효소연쇄반응 § 주형 DNA 분자의 특정 부분을 선택적으로 증폭 Primer set와 내열성 DNA polymerase를 이용하여 짧은 시간 내에 미량의 시료에서 § Kary Mullis 1983 (1985) 1993, Nobel Prize in Chemistry

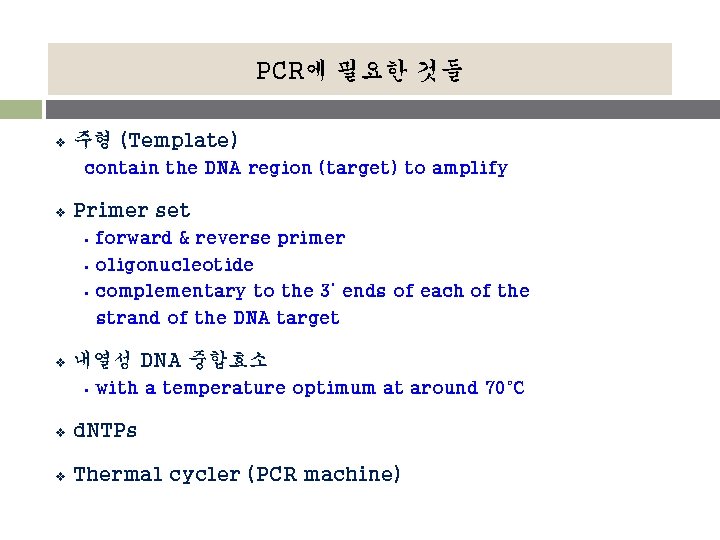
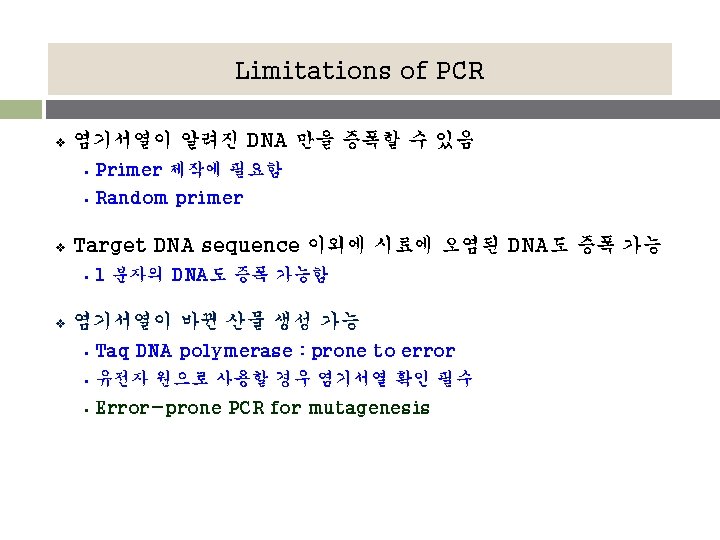
PCR에 필요한 것들 v 주형 (Template) contain the DNA region (target) to amplify v Primer set § § § v forward & reverse primer oligonucleotide complementary to the 3’ ends of each of the strand of the DNA target 내열성 DNA 중합효소 § with a temperature optimum at around 70°C v d. NTPs v Thermal cycler (PCR machine)

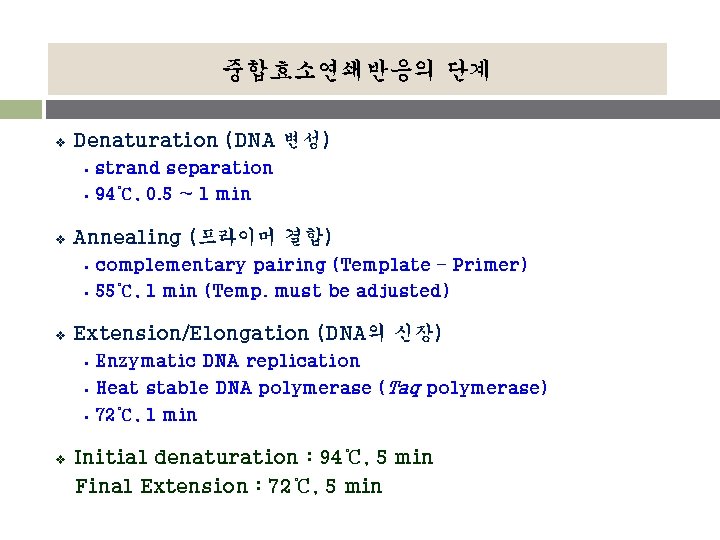
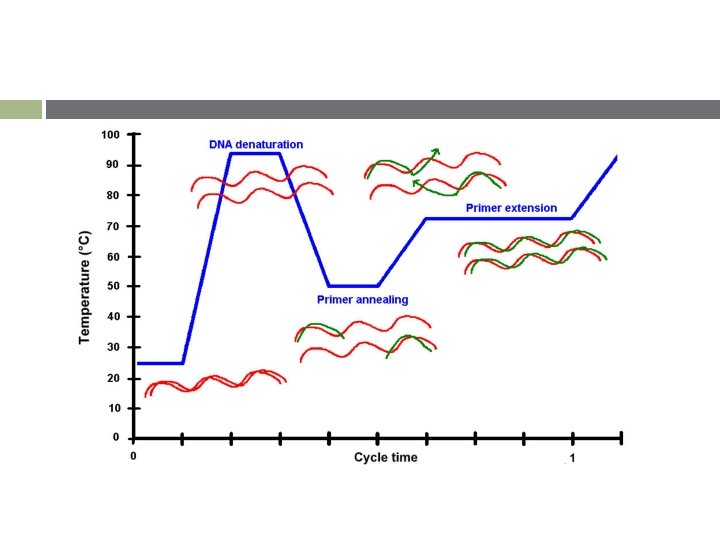
중합효소연쇄반응의 단계 v Denaturation (DNA 변성) § § v Annealing (프라이머 결합) § § v complementary pairing (Template – Primer) 55℃, 1 min (Temp. must be adjusted) Extension/Elongation (DNA의 신장) § § § v strand separation 94℃, 0. 5 ~ 1 min Enzymatic DNA replication Heat stable DNA polymerase (Taq polymerase) 72℃, 1 min Initial denaturation : 94℃, 5 min Final Extension : 72℃, 5 min


Thermal cycler For repeated heating and cooling

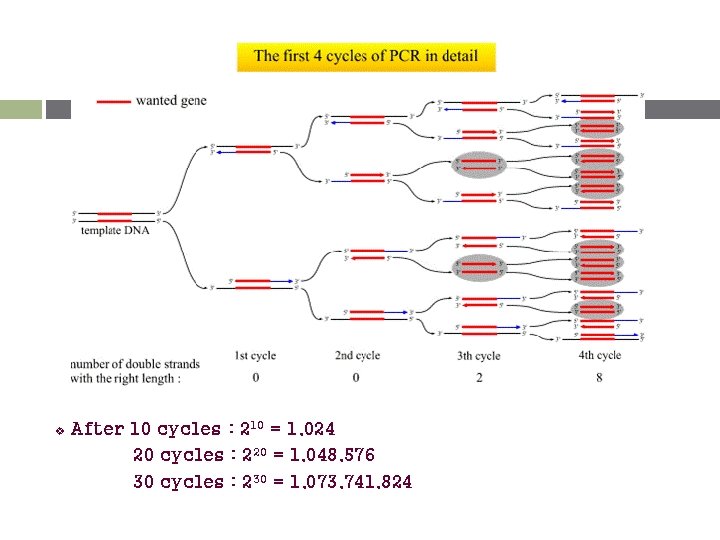
v After 10 cycles : 210 = 1, 024 20 cycles : 220 = 1, 048, 576 30 cycles : 230 = 1, 073, 741, 824

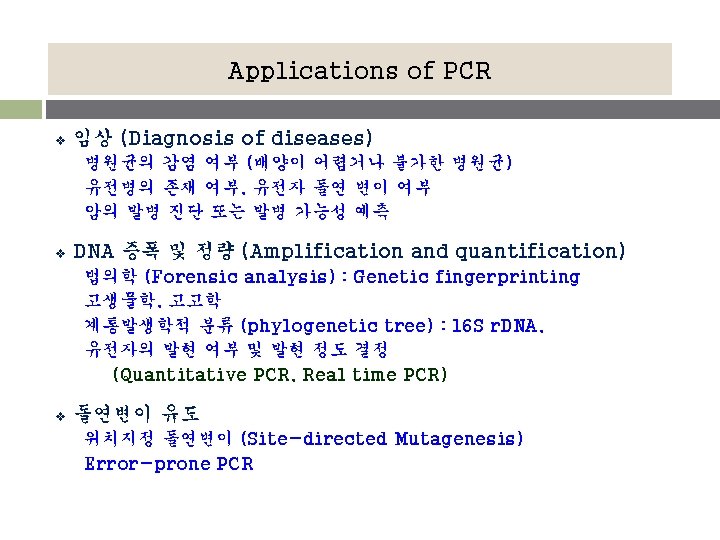
Applications of PCR v 임상 (Diagnosis of diseases) 병원균의 감염 여부 (배양이 어렵거나 불가한 병원균) 유전병의 존재 여부, 유전자 돌연 변이 여부 암의 발병 진단 또는 발병 가능성 예측 v DNA 증폭 및 정량 (Amplification and quantification) 법의학 (Forensic analysis) : Genetic fingerprinting 고생물학, 고고학 계통발생학적 분류 (phylogenetic tree) : 16 S r. DNA, 유전자의 발현 여부 및 발현 정도 결정 (Quantitative PCR, Real time PCR) v 돌연변이 유도 위치지정 돌연변이 (Site-directed Mutagenesis) Error-prone PCR


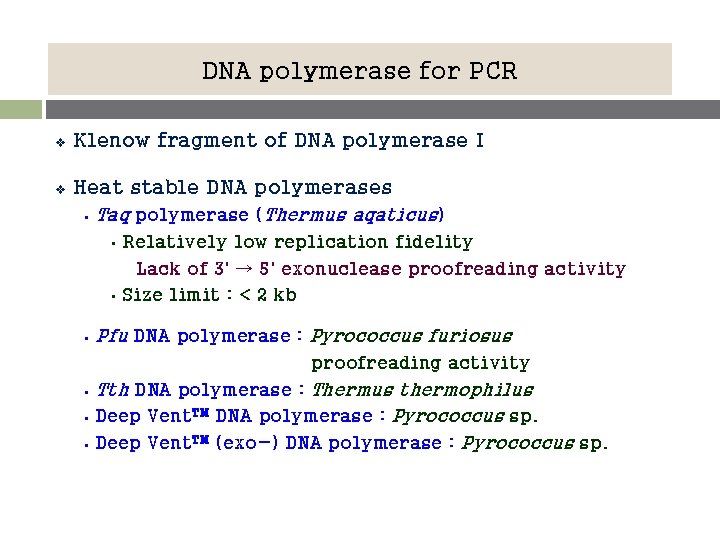
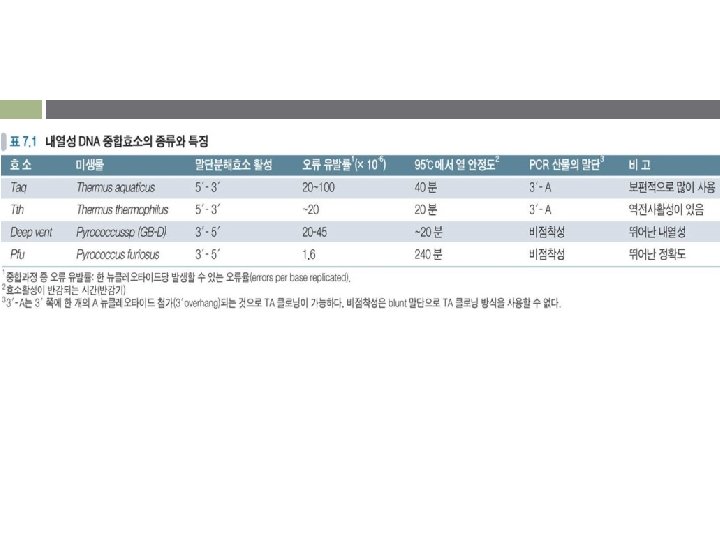
DNA polymerase for PCR v Klenow fragment of DNA polymerase I v Heat stable DNA polymerases § Taq polymerase (Thermus aqaticus) • • § Relatively low replication fidelity Lack of 3' → 5' exonuclease proofreading activity Size limit : < 2 kb Pfu DNA polymerase : Pyrococcus furiosus proofreading activity § § § Tth DNA polymerase : Thermus thermophilus Deep Vent. TM DNA polymerase : Pyrococcus sp. Deep Vent. TM (exo-) DNA polymerase : Pyrococcus sp.


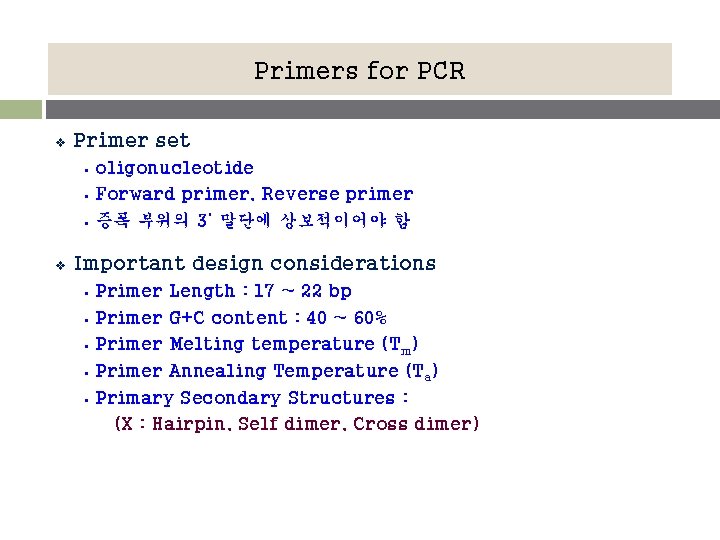
Primers for PCR v Primer set § § § v oligonucleotide Forward primer, Reverse primer 증폭 부위의 3’ 말단에 상보적이어야 함 Important design considerations § § § Primer Length : 17 ~ 22 bp Primer G+C content : 40 ~ 60% Primer Melting temperature (Tm) Primer Annealing Temperature (Ta) Primary Secondary Structures : (X : Hairpin, Self dimer, Cross dimer)

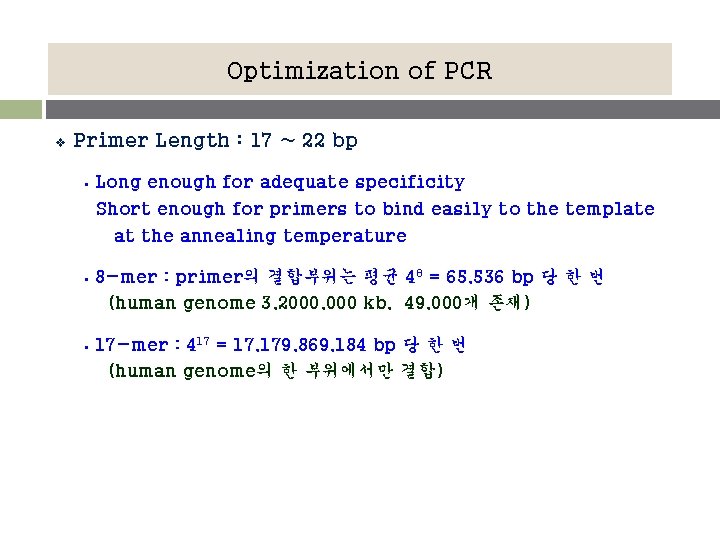
Optimization of PCR v Primer Length : 17 ~ 22 bp § Long enough for adequate specificity Short enough for primers to bind easily to the template at the annealing temperature § 8 -mer : primer의 결합부위는 평균 48 = 65, 536 bp 당 한 번 (human genome 3, 2000, 000 kb, 49, 000개 존재) § 17 -mer : 417 = 17, 179, 869, 184 bp 당 한 번 (human genome의 한 부위에서만 결합)

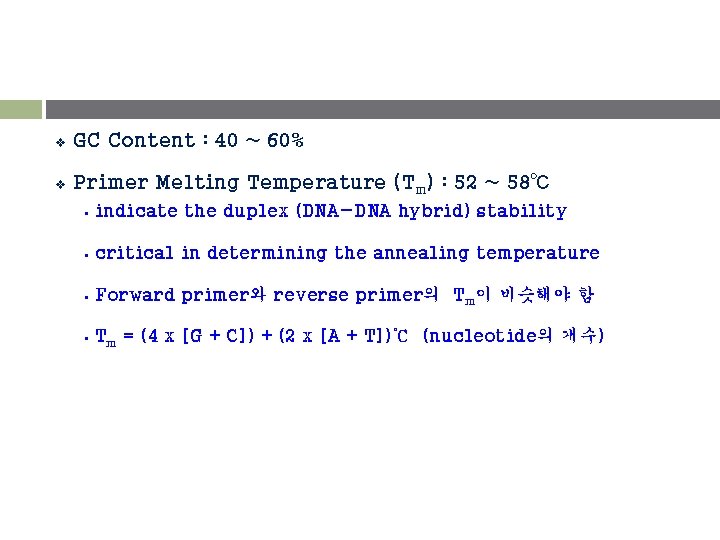
v GC Content : 40 ~ 60% v Primer Melting Temperature (Tm) : 52 ~ 58℃ § indicate the duplex (DNA-DNA hybrid) stability § critical in determining the annealing temperature § Forward primer와 reverse primer의 Tm이 비슷해야 함 § Tm = (4 x [G + C]) + (2 x [A + T])℃ (nucleotide의 개수)

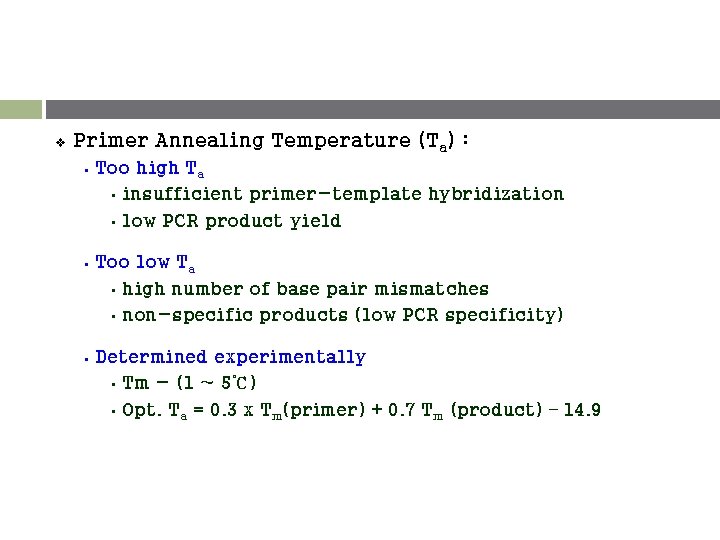
v Primer Annealing Temperature (Ta) : § Too high Ta • insufficient primer-template hybridization • low PCR product yield § Too low Ta • high number of base pair mismatches • non-specific products (low PCR specificity) § Determined experimentally • Tm - (1 ~ 5℃) • Opt. Ta = 0. 3 x Tm(primer) + 0. 7 Tm (product) – 14. 9

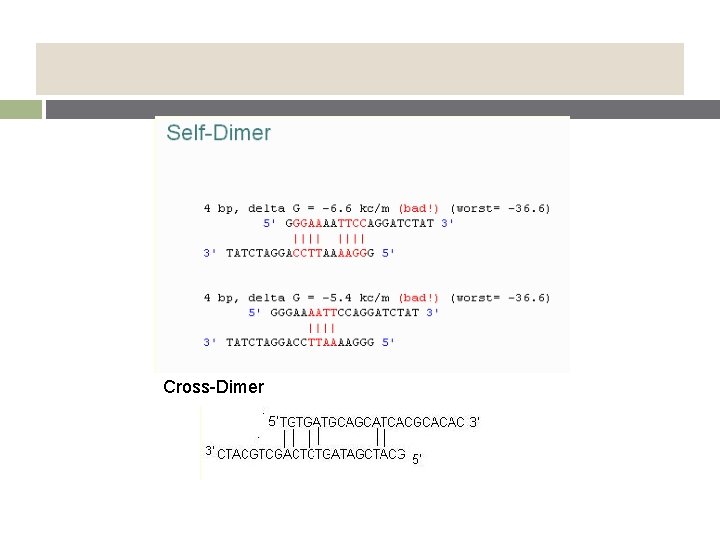
v Primer Secondary Structures § by intermolecular or intramolecular interactions § Hairpin : intramolecular Self dimer : intermolecular, F-F, R-R Cross dimer : intermolecular, F-R § adversely affect primer template annealing can lead to poor or no yield of the product


Cross-Dimer

v Length of the Target 0. 1 kb ~ 3 kb (~ 1 kb) ~ 10 kb, ~ 40 kb v Buffer solution v Bivalent cations : generally Mg 2+ is used Mn 2+ can be used for PCR-mediated DNA mutagenesis v Monovalent cation potassium ion (K+)

http: //www. youtube. com/watch? v=Zmqq. RPISg 0 g

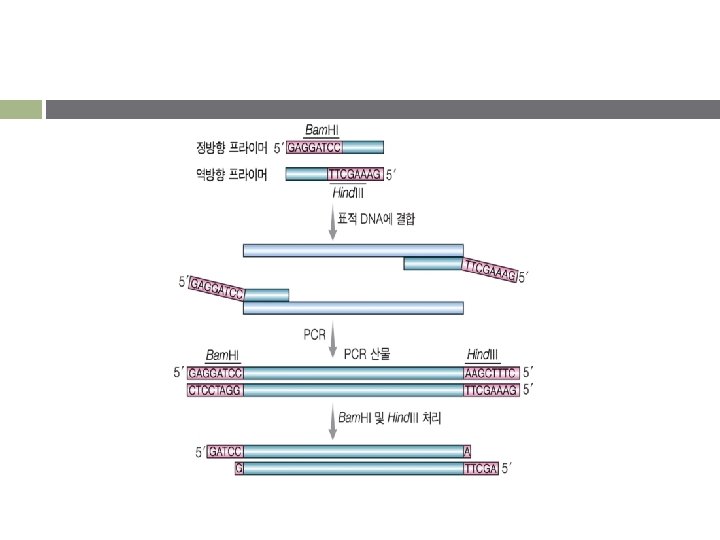
Cloning of PCR products v TA cloning § Taq, Tth : PCR product의 3’- 말단에 A 첨가 § v 제한효소 자리가 있는 primer를 사용하는 방법 § § v T vector 사용 Primer의 5’ 방향에 restriction site를 가진 oligonucletide가 결합된 primer PCR product를 제한효소로 처리 후 vector에 삽입 Topoisomerase와 연결된 vector를 이용하는 방법
