Plant viruses Plant viruses Nucleic acid in a





- Slides: 25

Plant viruses

Plant viruses • Nucleic acid in a protein capsid – Protein capsid • protection of NA • transfer (infection) • no membrane envelop – Nucleic acid • different types • infectious itself only in some viruses • encode just few genes (x bacteriophages up to 70) • Most enzymatic activities necessary for virus replication provided by the host cell

Viral genome - highly compact – HOW to save space? – formation of polyproteins – overlapping reading frames: alt. translation starts (transcription from both strands) - varying arrangement and strategies of expression – alt. read-through stop codon (translational readthrough) – alternative frameshift during translation – IRES (cap independent initiation of translation) – segmented genome (alt. more virions - e. g. Tobacco rattle virus)

Proteins encoded by plant viruses • Capsid proteins • Polymerases of NA • Movement proteins - transport through plasmodesmata • Suppressors of silencing Present in most plant viruses • Other specialized proteins: – e. g. proteases (cleavage of polyproteins), modulators of cell cycle, replication protein, helicases…. Varying representation of these proteins in plant viruses

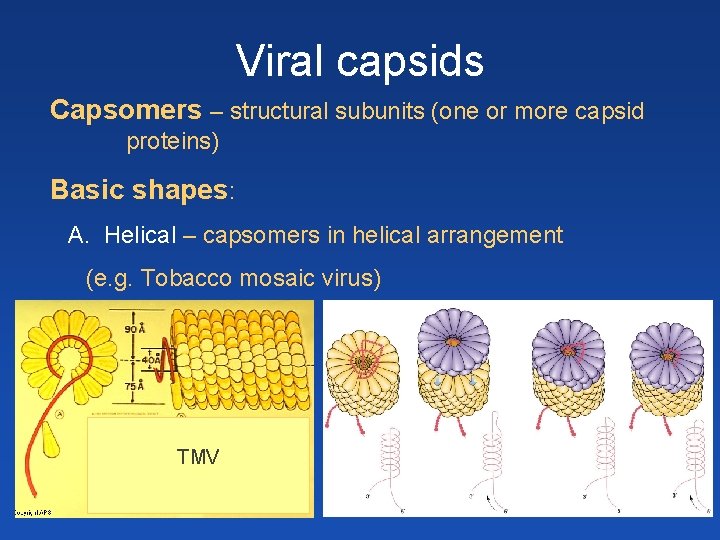
Viral capsids Capsomers – structural subunits (one or more capsid proteins) Basic shapes: A. Helical – capsomers in helical arrangement (e. g. Tobacco mosaic virus) TMV

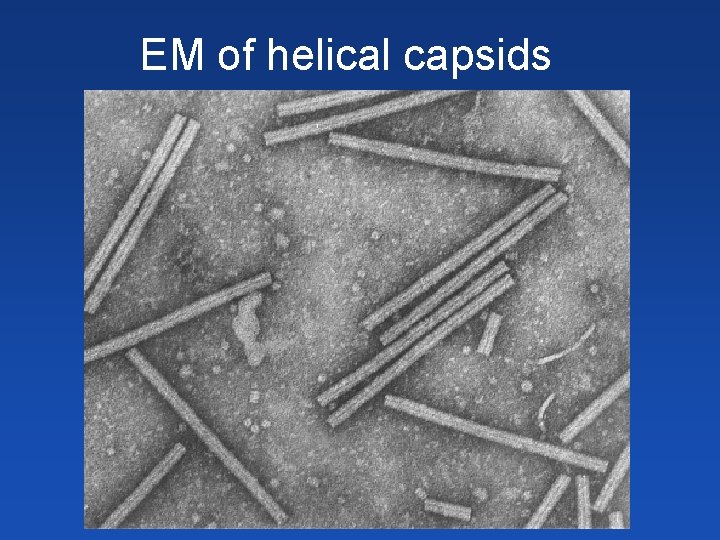
EM of helical capsids

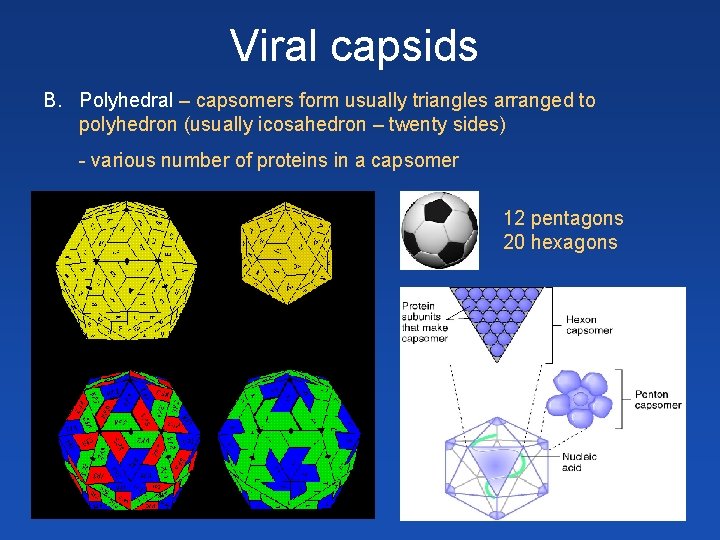
Viral capsids B. Polyhedral – capsomers form usually triangles arranged to polyhedron (usually icosahedron – twenty sides) - various number of proteins in a capsomer 12 pentagons 20 hexagons

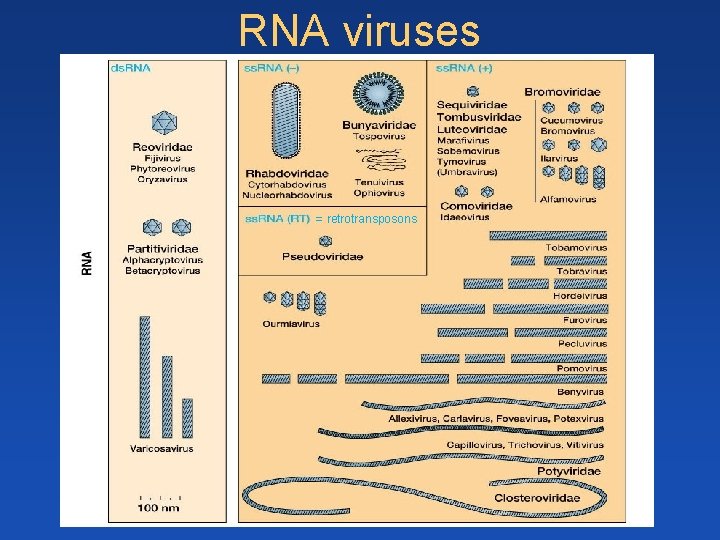
Classification of plant viruses - genome/mode of replication • ss. DNA • ds. DNA (RT) • ss. RNA • ds. RNA ss. RNA(+) = coding ss. RNA(-) = non-coding

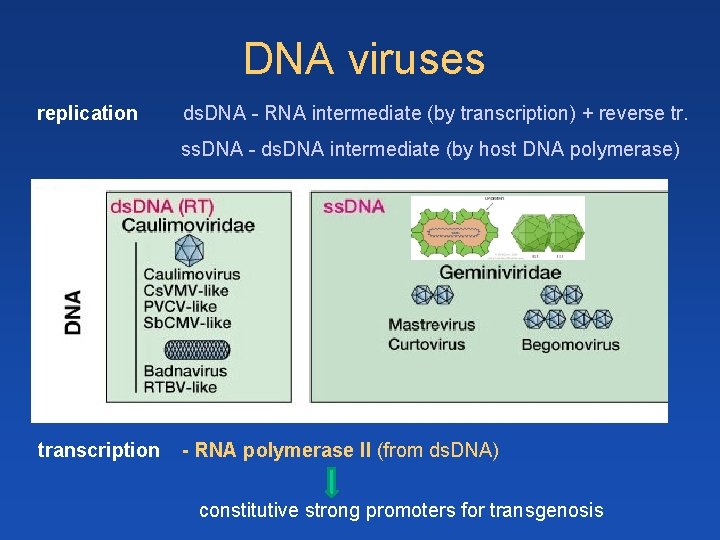
DNA viruses replication ds. DNA - RNA intermediate (by transcription) + reverse tr. ss. DNA - ds. DNA intermediate (by host DNA polymerase) transcription - RNA polymerase II (from ds. DNA) constitutive strong promoters for transgenosis

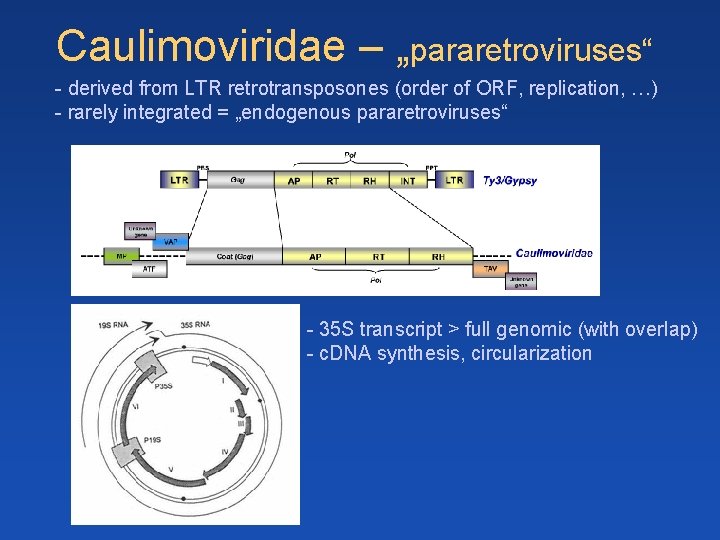
Caulimoviridae – „pararetroviruses“ - derived from LTR retrotransposones (order of ORF, replication, …) - rarely integrated = „endogenous pararetroviruses“ - 35 S transcript > full genomic (with overlap) - c. DNA synthesis, circularization

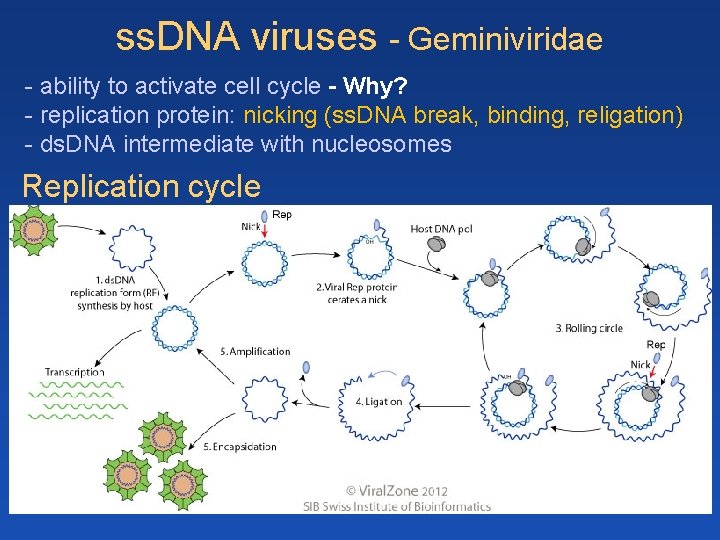
ss. DNA viruses - Geminiviridae - ability to activate cell cycle - Why? - replication protein: nicking (ss. DNA break, binding, religation) - ds. DNA intermediate with nucleosomes Replication cycle

RNA viruses = retrotransposons

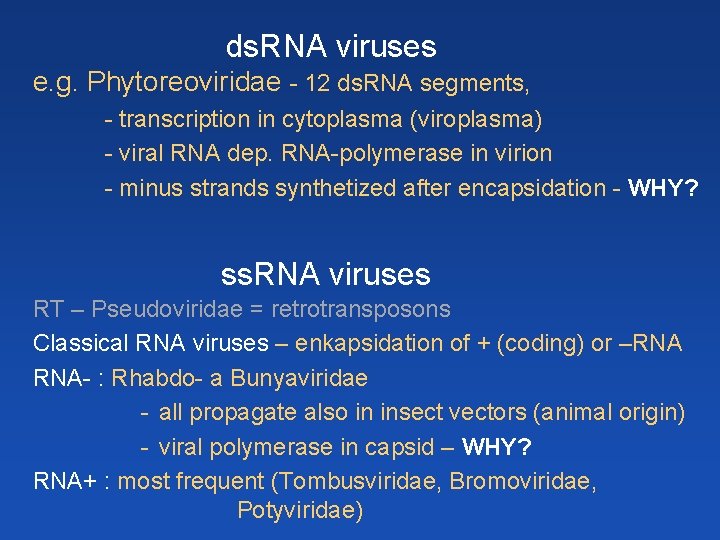
ds. RNA viruses e. g. Phytoreoviridae - 12 ds. RNA segments, - transcription in cytoplasma (viroplasma) - viral RNA dep. RNA-polymerase in virion - minus strands synthetized after encapsidation - WHY? ss. RNA viruses RT – Pseudoviridae = retrotransposons Classical RNA viruses – enkapsidation of + (coding) or –RNA RNA- : Rhabdo- a Bunyaviridae - all propagate also in insect vectors (animal origin) - viral polymerase in capsid – WHY? RNA+ : most frequent (Tombusviridae, Bromoviridae, Potyviridae)

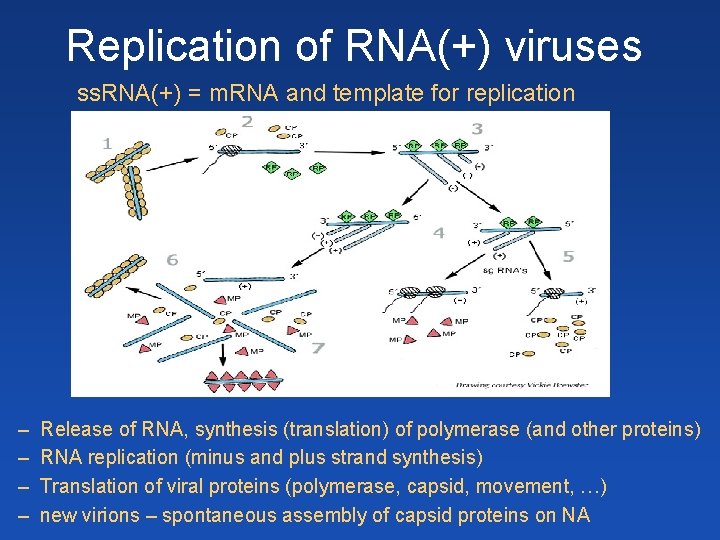
Replication of RNA(+) viruses ss. RNA(+) = m. RNA and template for replication – – Release of RNA, synthesis (translation) of polymerase (and other proteins) RNA replication (minus and plus strand synthesis) Translation of viral proteins (polymerase, capsid, movement, …) new virions – spontaneous assembly of capsid proteins on NA

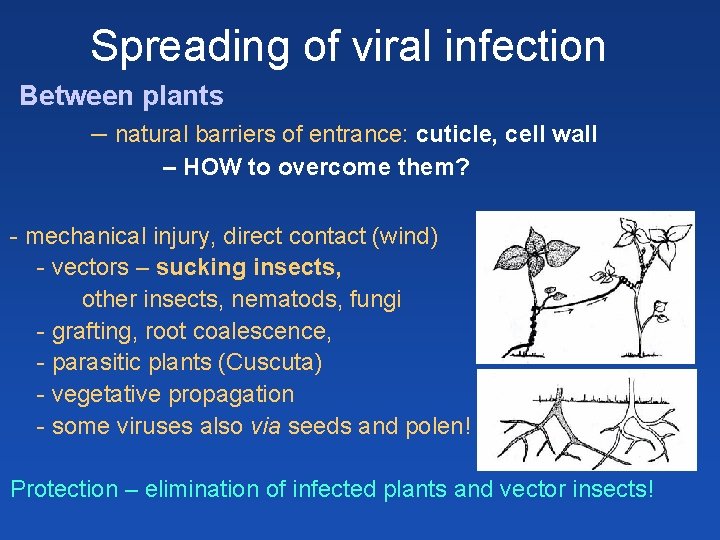
Spreading of viral infection Between plants – natural barriers of entrance: cuticle, cell wall – HOW to overcome them? - mechanical injury, direct contact (wind) - vectors – sucking insects, other insects, nematods, fungi - grafting, root coalescence, - parasitic plants (Cuscuta) - vegetative propagation - some viruses also via seeds and polen! Protection – elimination of infected plants and vector insects!

Transmission via sucking insects Non-persistant • adsorbtion on styletes (specific binding sites on acrostyle) • only transient infectiousness: immediate, persists minutes to hours Circulative • circulation of virus in insect body – salivary glands • infectiousness: latent period (hours to days), gradually decreasing many days Propagative • virus replication in transmittor insect • infectiousness: latent period (hours to days), life-long (also transmission to progeny)

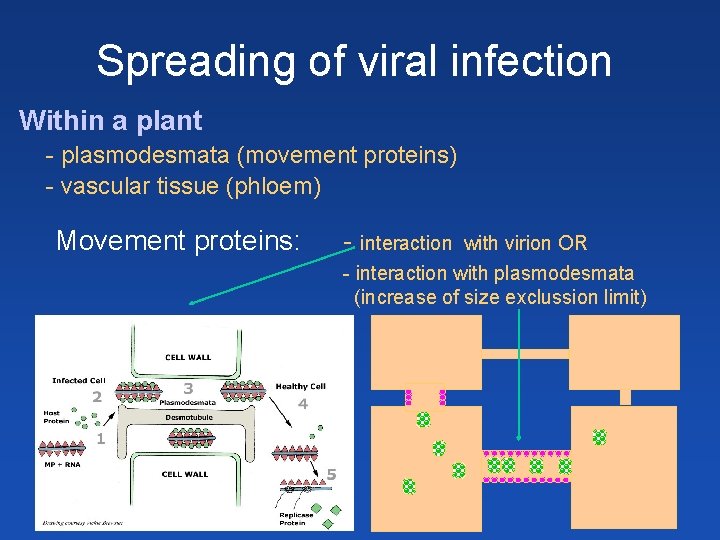
Spreading of viral infection Within a plant - plasmodesmata (movement proteins) - vascular tissue (phloem) Movement proteins: - interaction with virion OR - interaction with plasmodesmata (increase of size exclussion limit)

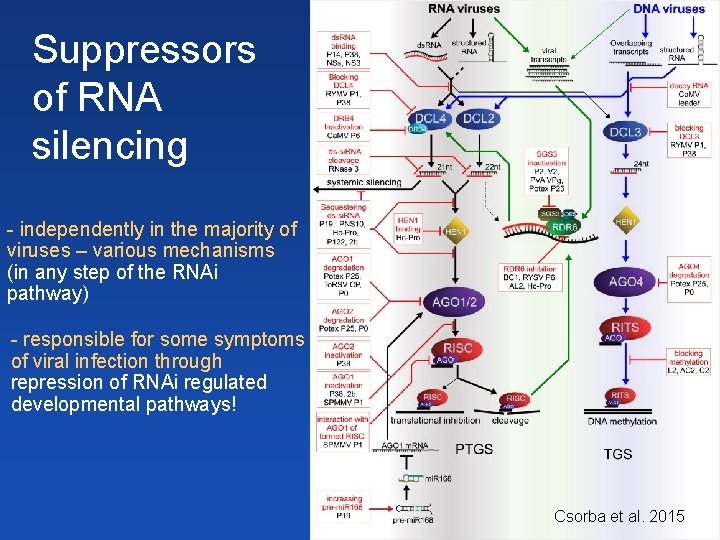
Suppressors of RNA silencing - independently in the majority of viruses – various mechanisms (in any step of the RNAi pathway) - responsible for some symptoms of viral infection through repression of RNAi regulated developmental pathways! Csorba et al. 2015

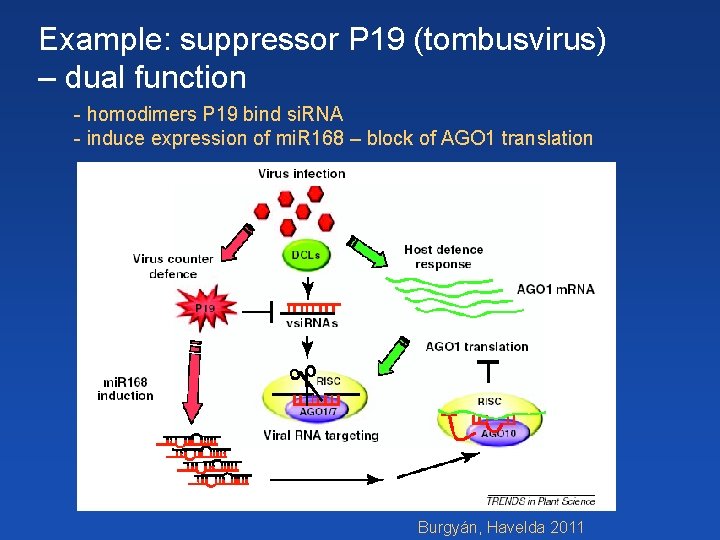
Example: suppressor P 19 (tombusvirus) – dual function - homodimers P 19 bind si. RNA - induce expression of mi. R 168 – block of AGO 1 translation Burgyán, Havelda 2011

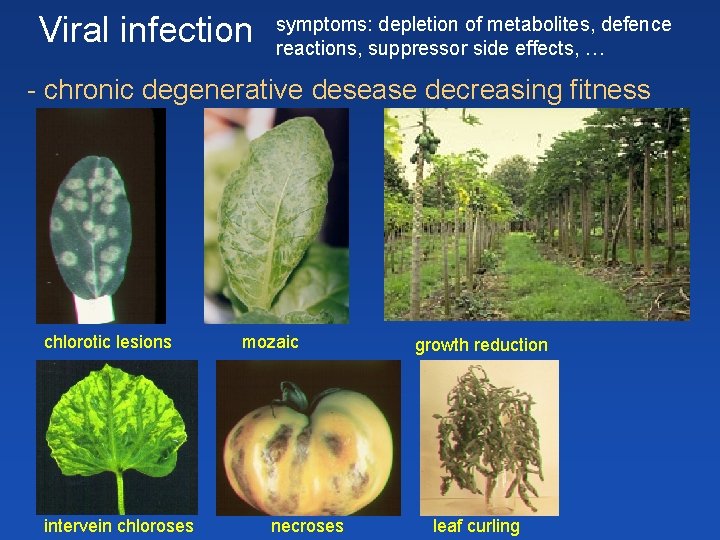
Viral infection symptoms: depletion of metabolites, defence reactions, suppressor side effects, … - chronic degenerative desease decreasing fitness chlorotic lesions intervein chloroses mozaic necroses growth reduction leaf curling

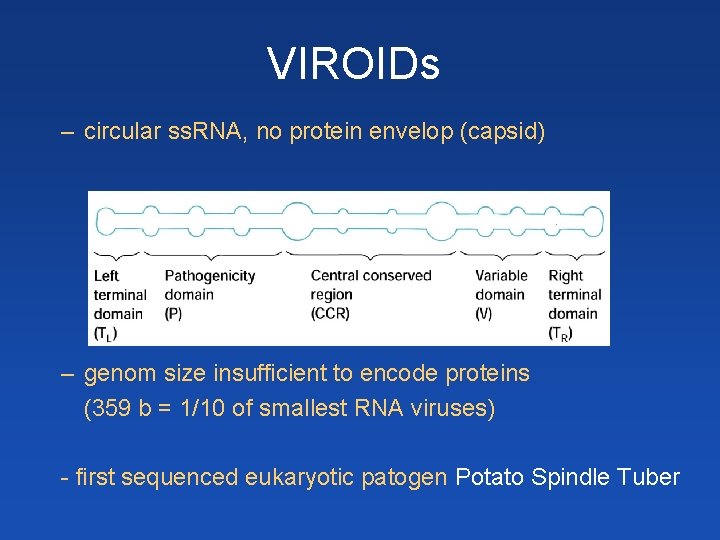
VIROIDs – circular ss. RNA, no protein envelop (capsid) – genom size insufficient to encode proteins (359 b = 1/10 of smallest RNA viruses) - first sequenced eukaryotic patogen Potato Spindle Tuber

VIROIDs Replication with host DNA dep. (!) RNA Pol II - probably rolling circle - concatemers of some viroids autocatalytically cleaved by hammer-head ribozyme - symptoms of infection – likely result from induced RNAi non-specifically affecting expression of plant genes - common features (origin? ) with HDV (hepatitis D virus)


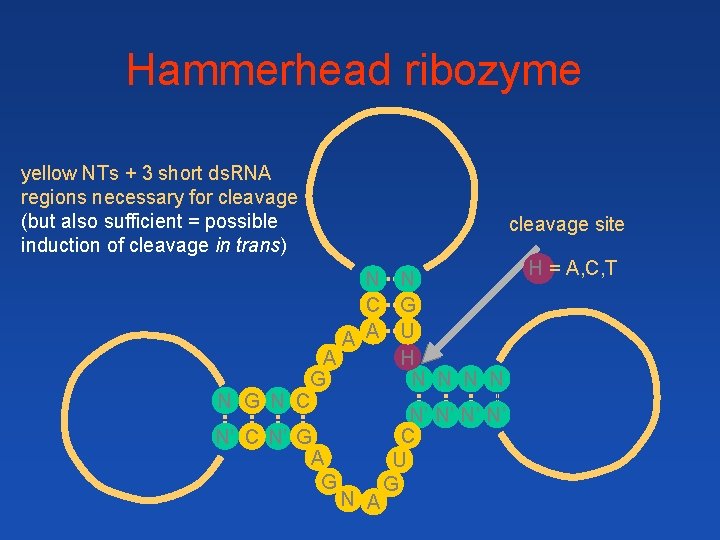
Hammerhead ribozyme yellow NTs + 3 short ds. RNA regions necessary for cleavage (but also sufficient = possible induction of cleavage in trans) N N C G A A U A H G N N N G N C N’ N’ C N’ G A U G G N A cleavage site H = A, C, T

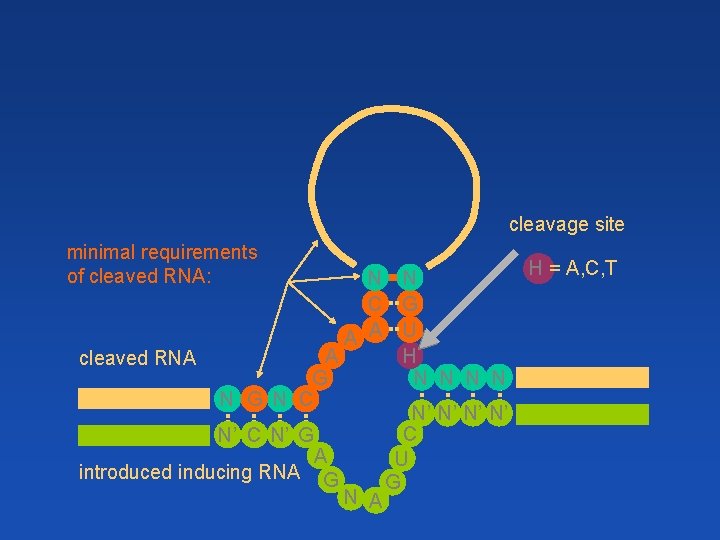
cleavage site minimal requirements of cleaved RNA: N N C G A A U A H cleaved RNA G N N N G N C N’ N’ C N’ G A U introduced inducing RNA G G N A H = A, C, T