Plant Genomes Houses of genetic materials Total genetic














- Slides: 49

Plant Genomes Houses of genetic materials Total genetic material within a cell Usually referred to a haploid cell ]Basic set of genetic material (1 x[(

Plant Genomes General perception: DNA in Nucleus Other organelles: Mitochondria / Chloroplast May not correlate with chromosome / gene number Extensive transfer of genes from organelles to nucleus

Plant Genomes

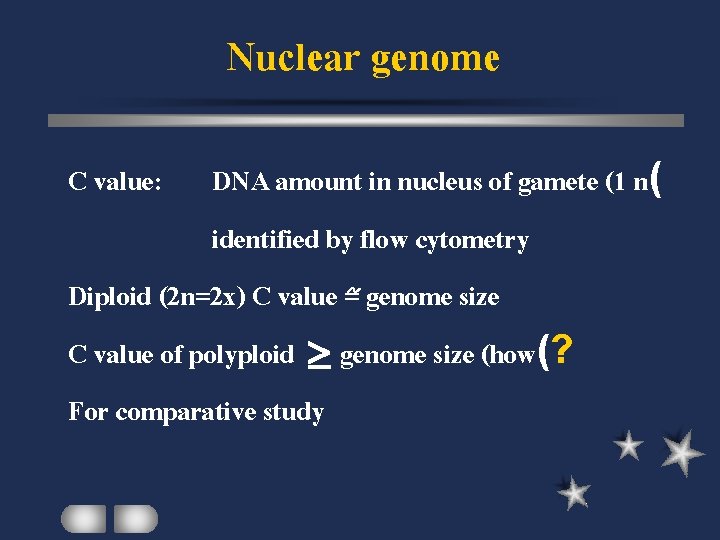
Nuclear genome C value: DNA amount in nucleus of gamete (1 n( identified by flow cytometry Diploid (2 n=2 x) C value = genome size C value of polyploid > genome size (how(? For comparative study

Typical C value bacteria 1. 5 * 107 mammals 1 -2 * 109 spore-bearing vascular plants gymnosperm 70 * 109 angiosperm 5 -30 * 109 bp bp 50 * 109 bp bp bp

Typical C value Highly varied C value in plant Plants in general with big genome Variation of DNA amounts among closely related plants

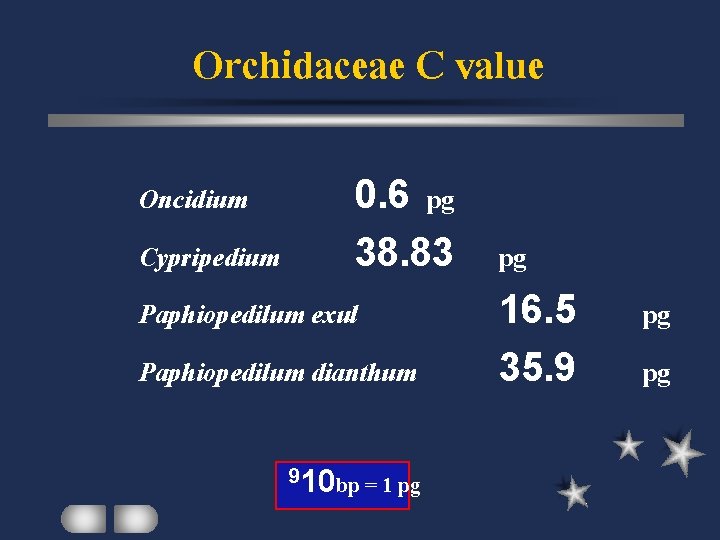
Orchidaceae C value Oncidium 0. 6 pg Cypripedium 38. 83 Paphiopedilum exul Paphiopedilum dianthum 910 bp = 1 pg pg 16. 5 35. 9 pg pg

Nuclear genome Increased genome size =Increased gene number Non-coding DNA regions: Variation in genome size ?

Nuclear genome Diversity: as a result of nt change 0. 1% every 2 * 105 years more diverse in non genic area why?

Gene evolution Major rearrangement by Recombination duplication or deletion or transpose Exons and Regulatory elements shuffled as separate modules resulting in new proteins / new roles

Gene evolution Minor change by Point Mutation following duplication replication errors of copies May result in related/unrelated gene products share common amino acid segments different functions

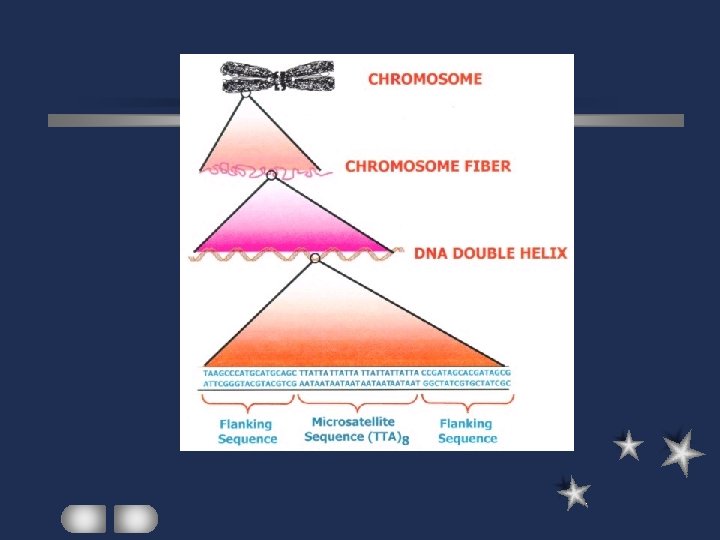
Composition of nuclear genome Unique sequence low copy number mostly genes Repetitive sequence satellite DNA transposable element

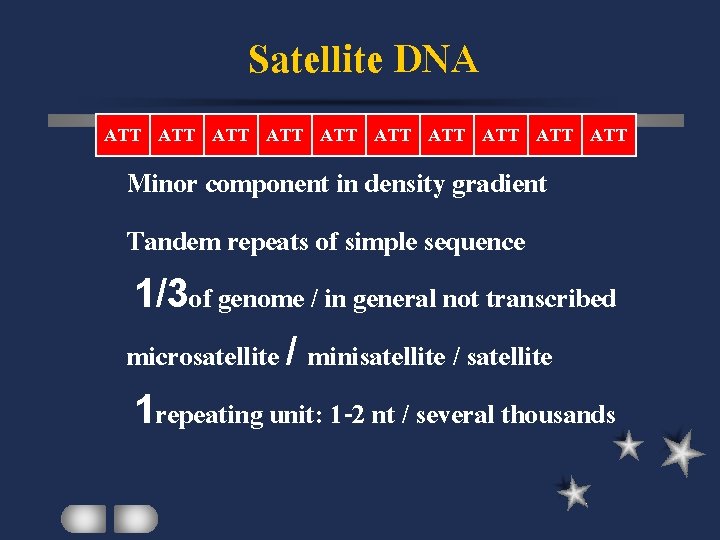
Satellite DNA ATT ATT ATT Minor component in density gradient Tandem repeats of simple sequence 1/3 of genome / in general not transcribed microsatellite / minisatellite / satellite 1 repeating unit: 1 -2 nt / several thousands

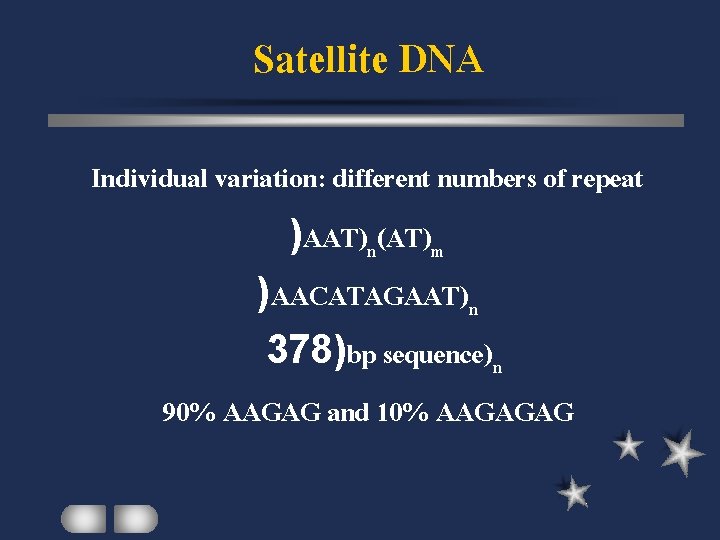
Satellite DNA Individual variation: different numbers of repeat )AAT)n(AT)m )AACATAGAAT)n 378)bp sequence)n 90% AAGAG and 10% AAGAGAG


Transposon Transposable element / Transposition / Transpose Movable segment of DNA : interspersed / scattered in nuclear genome DNA element: transposase RNA intermediate: reverse transcriptase

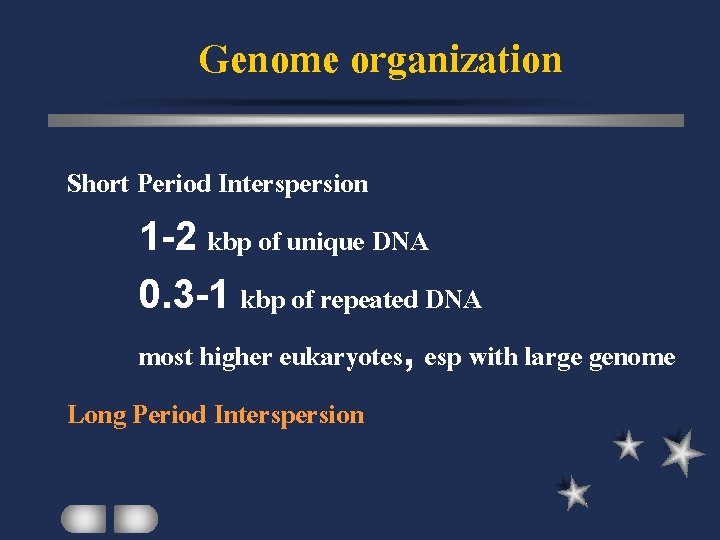
Genome organization Short Period Interspersion 1 -2 kbp of unique DNA 0. 3 -1 kbp of repeated DNA most higher eukaryotes, esp with large genome Long Period Interspersion

Genome organization Short Period Interspersion Long Period Interspersion alternated stretches of unique / repeated DNA longer than 10 kbp found in some eukaryotes with small genome size

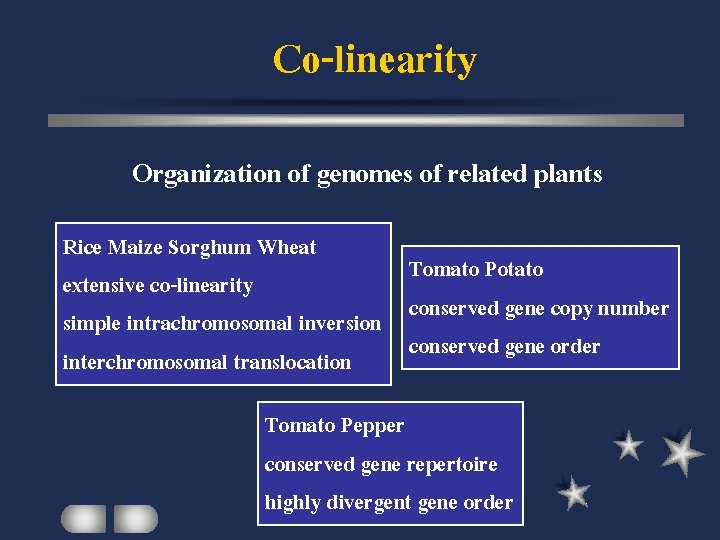
Co-linearity Organization of genomes of related plants Rice Maize Sorghum Wheat Tomato Potato extensive co-linearity conserved gene copy number simple intrachromosomal inversion conserved gene order interchromosomal translocation Tomato Pepper conserved gene repertoire highly divergent gene order

Organelle Genome Mitochondria / Chloroplast Small and simple DNA Usually circular Multiple copies Clusters in matrix / stroma

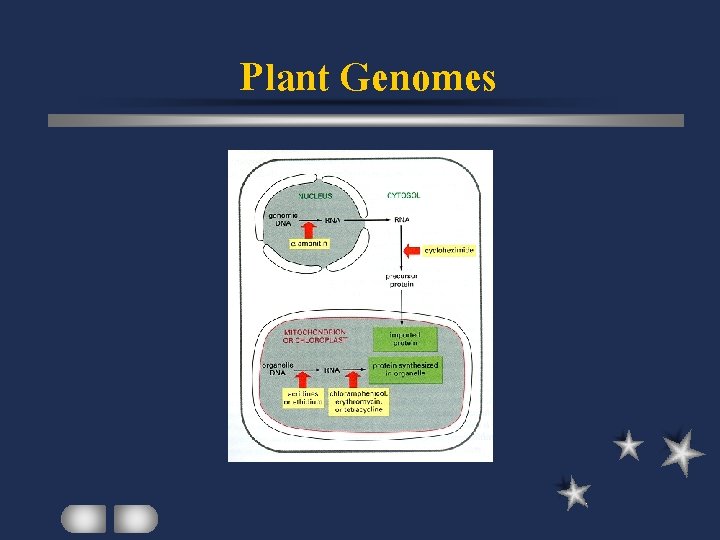
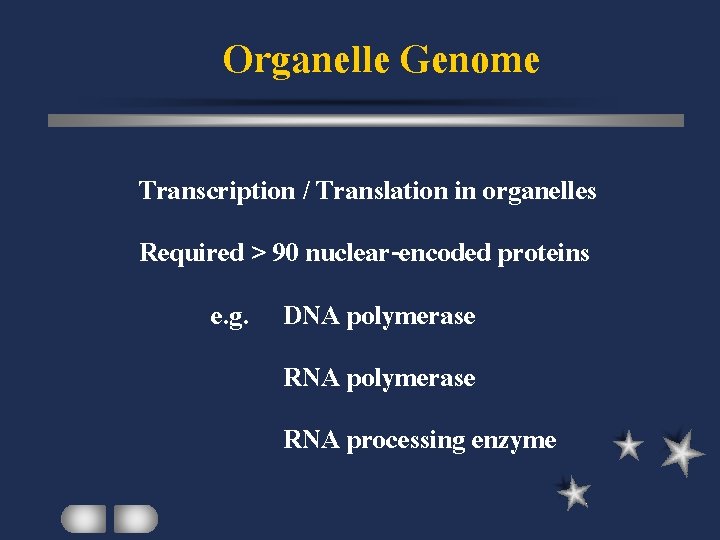
Organelle Genome Transcription / Translation in organelles Required > 90 nuclear-encoded proteins e. g. DNA polymerase RNA processing enzyme

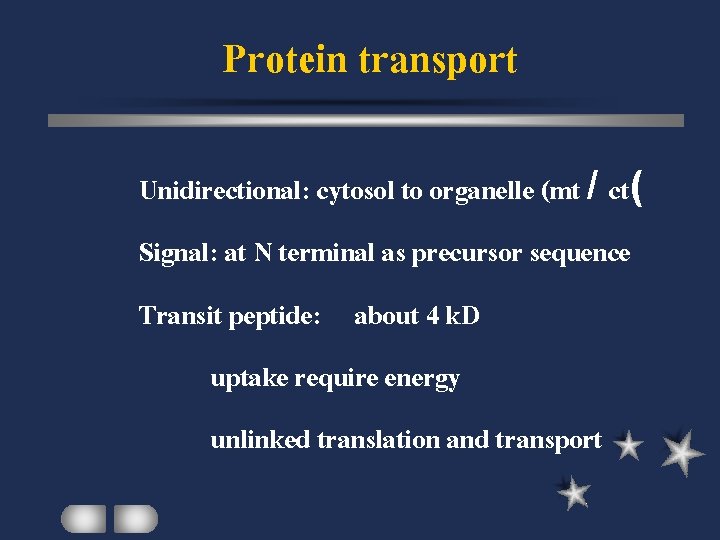
Protein transport Unidirectional: cytosol to organelle (mt / ct( Signal: at N terminal as precursor sequence Transit peptide: about 4 k. D uptake require energy unlinked translation and transport

Organelle Genome Isolated organelles Continue to make DNA / RNA / Protein for a brief period Complete genetic system

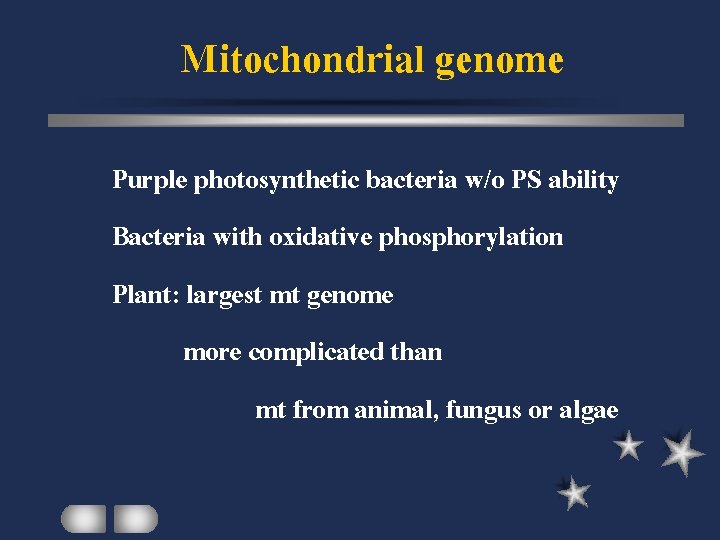
Mitochondrial genome Purple photosynthetic bacteria w/o PS ability Bacteria with oxidative phosphorylation Plant: largest mt genome more complicated than mt from animal, fungus or algae

Mitochondrial genome Small animal Fungus / Algae Plant 15 -16 kbp 20 -100 kbp 150 -2500 kbp

Mitochondrial genome Plant mt genome: large and variable but encode only a few more proteins Oenothera Turnip Corn Muskmelon 195 kbp 218 kbp 570 kbp 2400 kbp

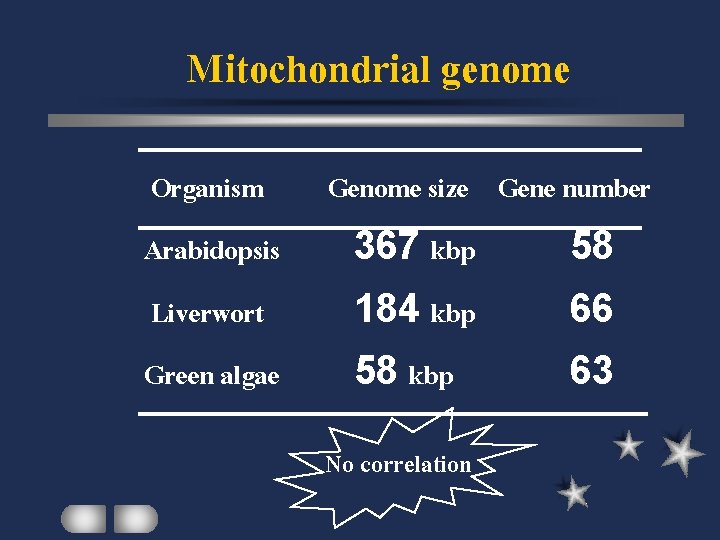
Mitochondrial genome Organism Genome size Gene number Arabidopsis 367 kbp 58 Liverwort 184 kbp 66 Green algae 58 kbp 63 No correlation

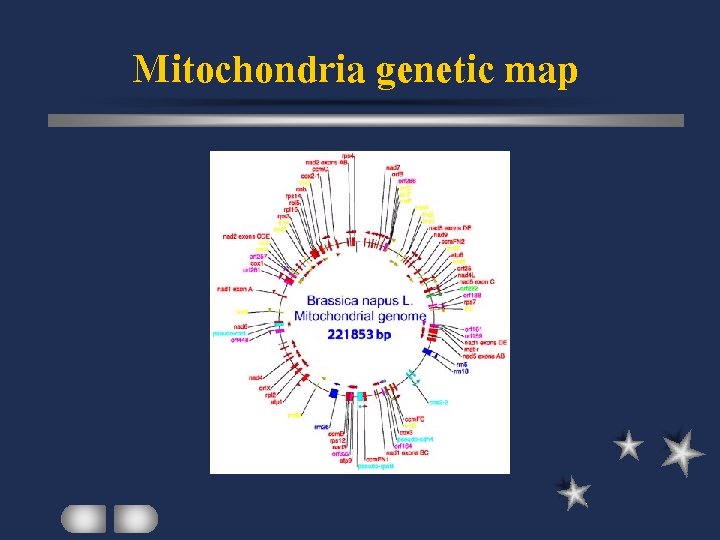
Plant mt genome Structure of plant mt genome Circular or linear or supercoiled Multipartite organization several molecules of different sizes Introns in some genes of some species

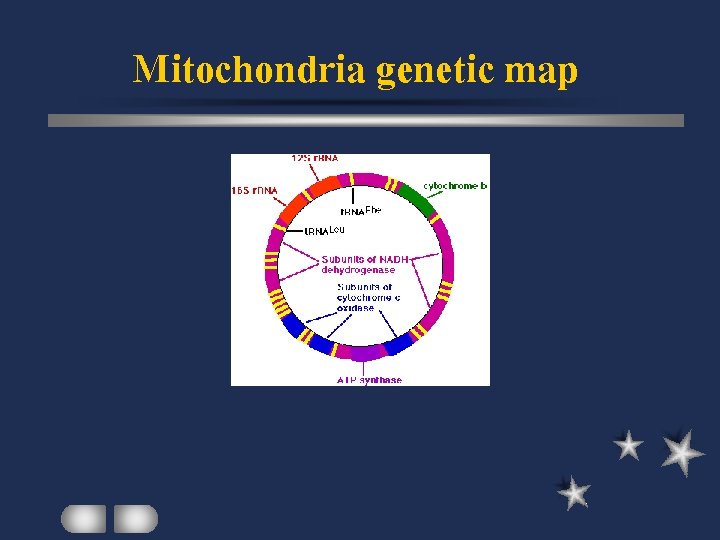
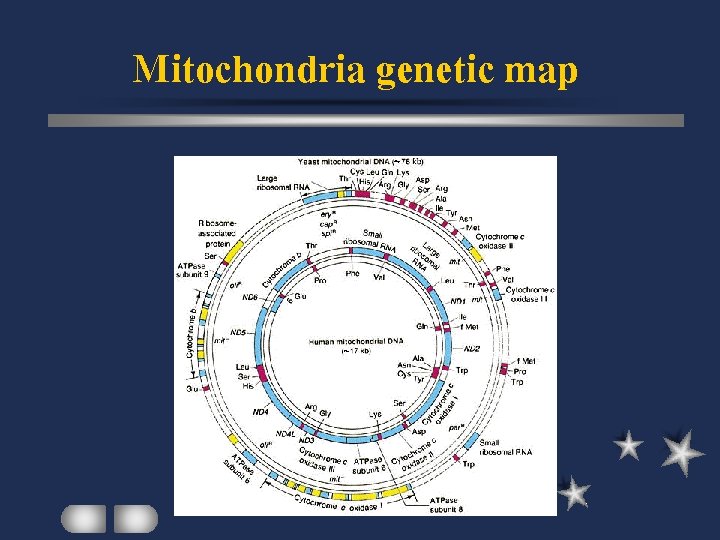
Plant mt gene product Genetic apparatus r. RNA, t. RNA, r. Protein Respiratory chain complex (energy metabolism( eg subunits I and II of Cytochrome c Oxidase ATPase subunits NADH dehydrogenase subunits

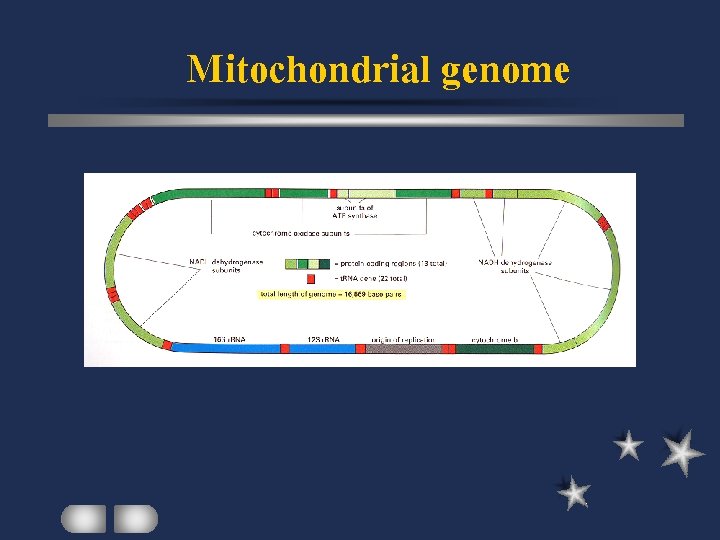
Mitochondrial genome

Mitochondria genetic map

Mitochondria genetic map

Mitochondria genetic map

Mitochondrial genes Cytoplasmic inheritance Maternal / Uniparental inheritance

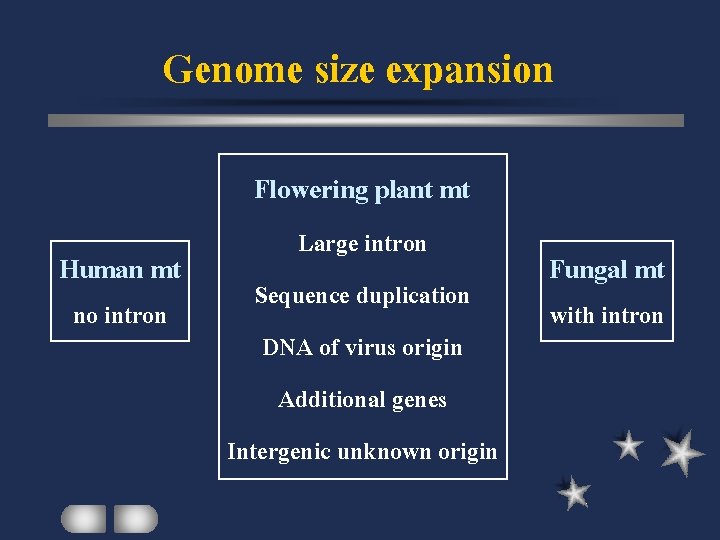
Genome size expansion Flowering plant mt Human mt no intron Large intron Sequence duplication DNA of virus origin Additional genes Intergenic unknown origin Fungal mt with intron

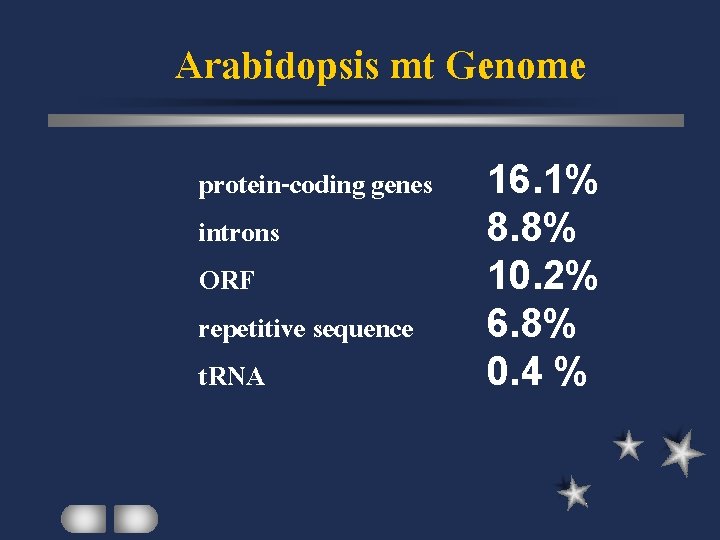
Arabidopsis mt Genome protein-coding genes introns ORF repetitive sequence t. RNA 16. 1% 8. 8% 10. 2% 6. 8% 0. 4 %

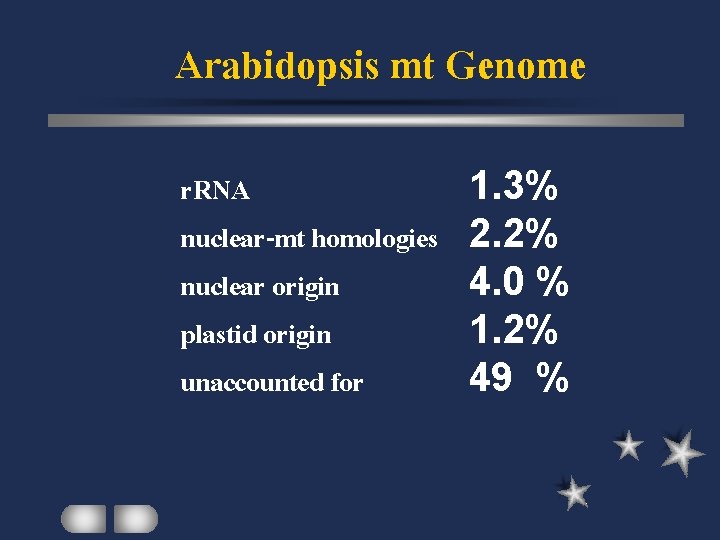
Arabidopsis mt Genome r. RNA nuclear-mt homologies nuclear origin plastid origin unaccounted for 1. 3% 2. 2% 4. 0 % 1. 2% 49 %

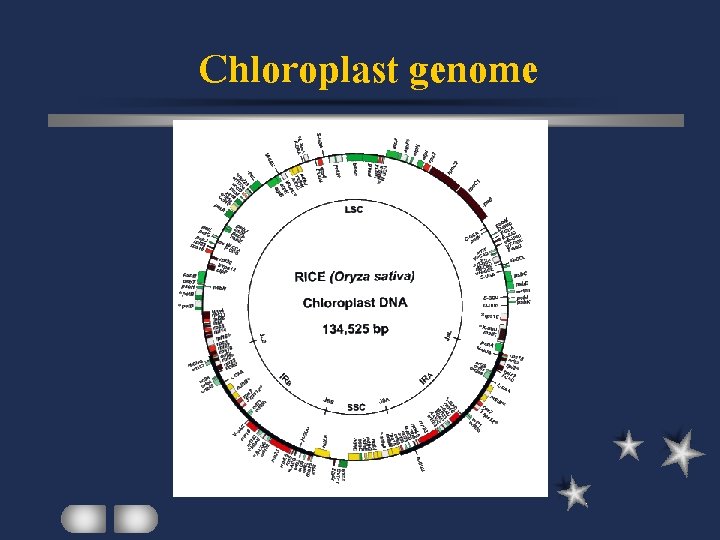
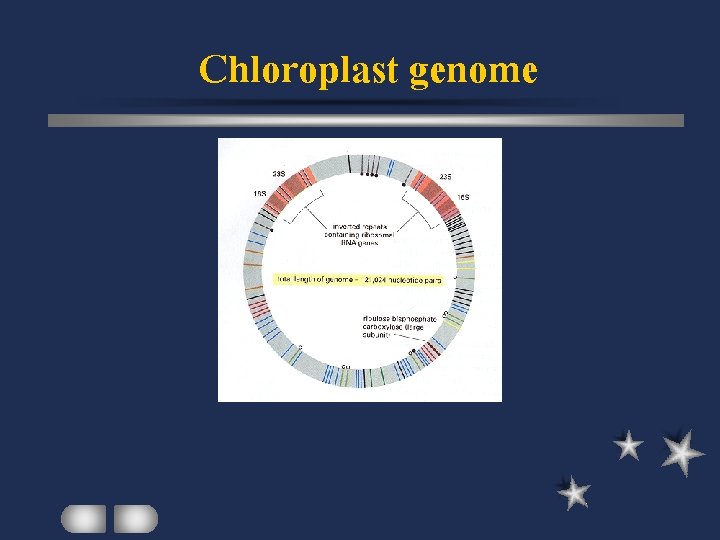
Chloroplast genome Photosynthetic cyanobacteria Similar sequence, size, structure, organization among organisms Differences in higher plant ct genomes accounted for by inverted repeats

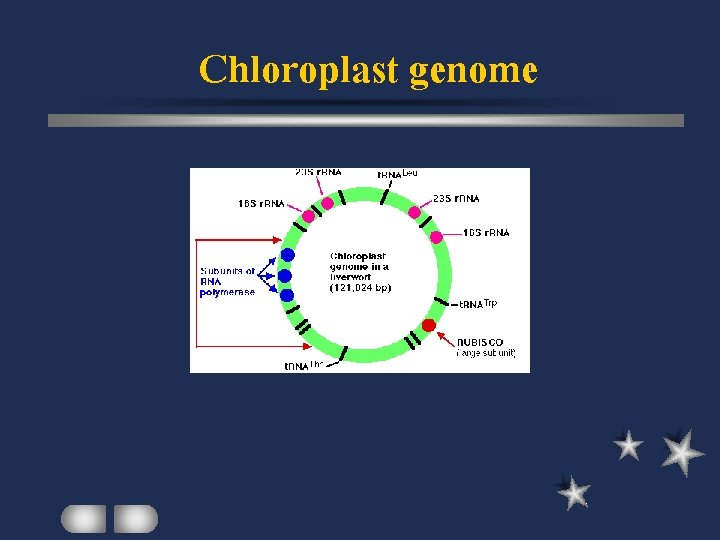
Chloroplast genome

Chloroplast genome

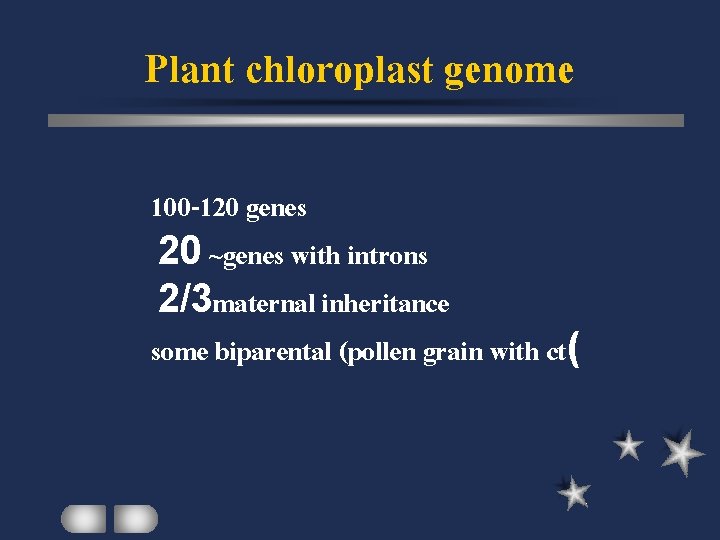
Plant chloroplast genome 100 -120 genes 20 ~genes with introns 2/3 maternal inheritance some biparental (pollen grain with ct(

Plant chloroplast genome Colinearity found in maize, petunia spinach, cucumber and mungbean Extensive rearrangement in pea and broadbean

Chloroplast Genome Chloroplast ribosome different from rbs in cytoplasm similar to rbs from E. coli structure / nt sequence antibiotic sensitivity

Chloroplast Genome Basic regulatory sequences )promoter / terminator( similar to bacterial genome

Chloroplast DNA Structure Circular or supercoiled About 125 -200 kbp Single copy / inverted repeat regions Low GC content (rich / poor regions( Most genes: polycistronic (except for rbc. L(

Chloroplast genome

Chloroplast genes Function: photosynthesis, fatty acid synthesis nitrogen metabolism and sulfate reduction Encode: Translation component t. RNA r. Protein Subunits of transcription Subunits of photosynthetic enzymes

Spinach ct genome Double-stranded circular 150 kbp 108 unique loci with know functions 17 genes with intron quadripartite

Spinach ct genome 2 inverted repeats of about 25 kbp 2 single-copy regions (large and small( RNA genes Protein genes