Phylogenetic analysis taken from http allserv rug ac






































- Slides: 80

Phylogenetic analysis taken from http: //allserv. rug. ac. be/~avierstr and http: //www. cs. otago. ac. nz/cosc 348/Lectur es/MSAPhylogeny. htm And Introduction to Bioinformatics course slides

Purpose of phylogenetics : • Reconstruct the evolutionary relationship between species Experience learns that closely related organisms have similar sequences, more distantly related organisms have more dissimilar sequences. • Estimate the time of divergence between two organisms since they last shared a common ancestor. But… • The theory and practical applications of the different models are not universally accepted. • Important to have a good alignment to start with. (Garbage in, Garbage out) • Trees based on an alignment of a gene represent the relationship between genes and this is not necessarily the same relationship as between the whole organisms. If trees are calculated based on different genes from organisms, it is possible that these trees result in different relationships.

Why is phylogeny imporant • Determining tree of life (e. g. , for a new organism) • Determining gene function • Understand which parts of the gene/regulatory sequences are important • Tracing the evolution of genes – horizontal gene transfer etc.

Protein or DNA? • As with Multiple Sequence Alignment – proteins are preferred – More informative – Shorter in length – Less chance of multiple mutations at the same site • When DNA? – A non-coding sequence – Proteins too similar

Terminology : • • • node : a node represents a taxonomic unit. This can be a taxon (an existing species) or an ancestor (unknown species : represents the ancestor of 2 or more species). branch : defines the relationship between the taxa in terms of descent and ancestry. topology : is the branching pattern. branch length : often represents the number of changes that have occurred in that branch. root : is the common ancestor of all taxa. distance scale : scale which represents the number of differences between sequences (e. g. 0. 1 means 10 % diff

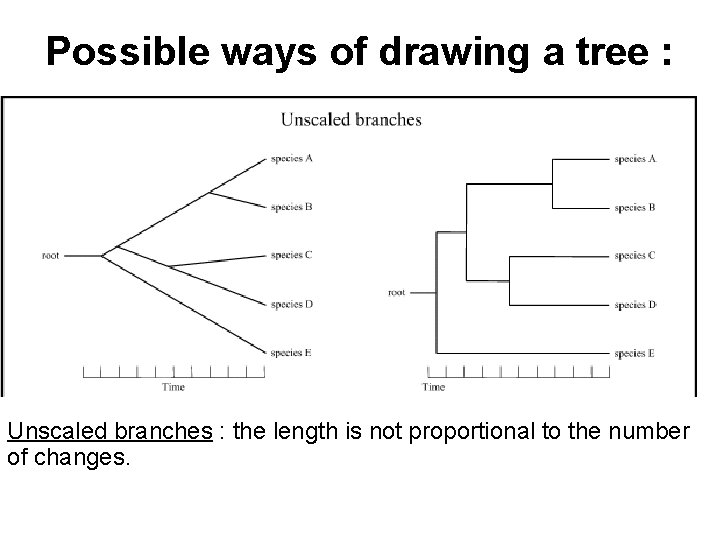
Possible ways of drawing a tree : Unscaled branches : the length is not proportional to the number of changes.

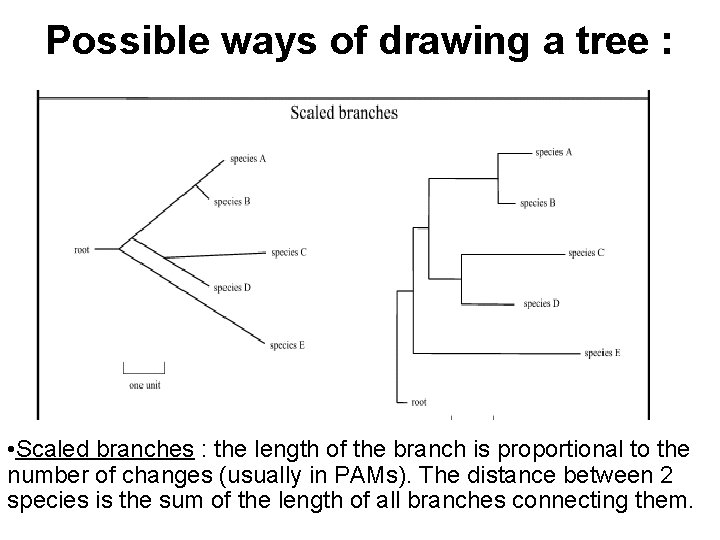
Possible ways of drawing a tree : • Scaled branches : the length of the branch is proportional to the number of changes (usually in PAMs). The distance between 2 species is the sum of the length of all branches connecting them.

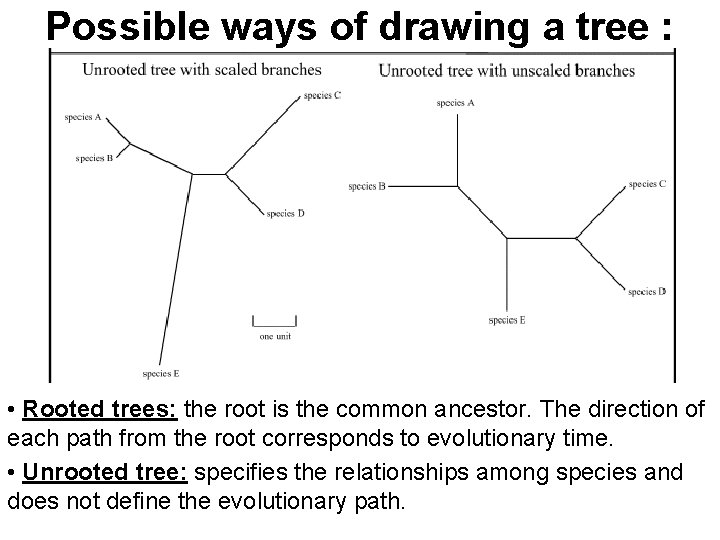
Possible ways of drawing a tree : • Rooted trees: the root is the common ancestor. The direction of each path from the root corresponds to evolutionary time. • Unrooted tree: specifies the relationships among species and does not define the evolutionary path.

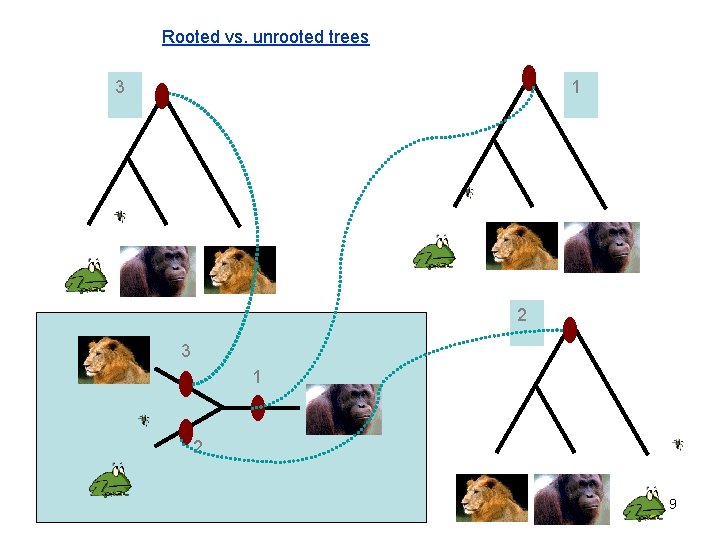
Rooted vs. unrooted trees 3 1 2 9

Rooted vs. Unrooted. The position of the root does not affect the MP score. 10

Intuition why rooting doesn’t change the score Gene number 1 1 or 0 1 s 1 s 4 s 3 s 2 s 5 1 1 1 0 0 The change will always be on the same branch, no matter where the root is positioned… 11

We want rooted trees! How can we root the tree? 12

13

14

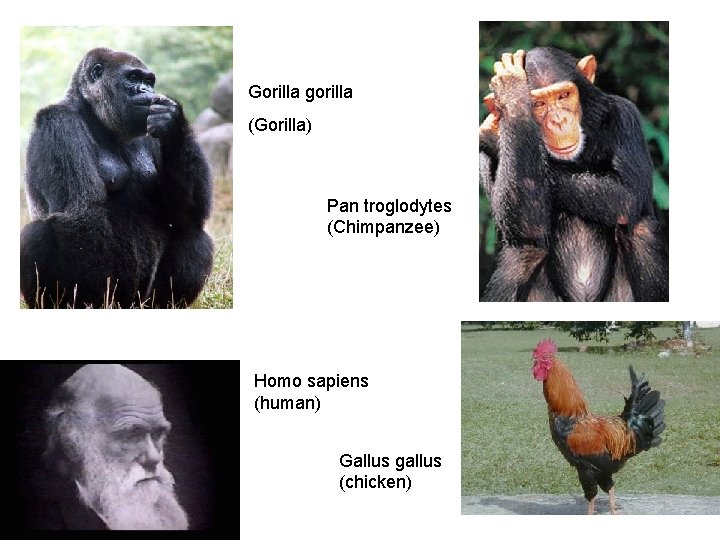
Gorilla gorilla (Gorilla) Pan troglodytes (Chimpanzee) Homo sapiens (human) Gallus gallus (chicken) 15

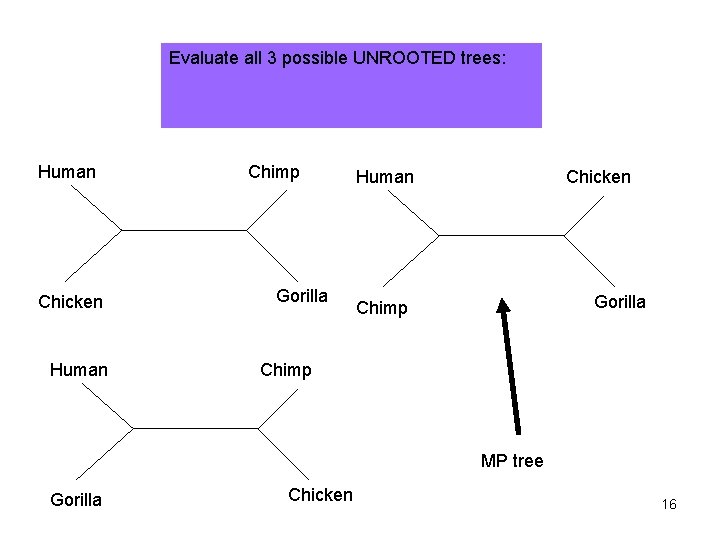
Evaluate all 3 possible UNROOTED trees: Human Chicken Human Chimp Gorilla Human Chicken Gorilla Chimp MP tree Gorilla Chicken 16

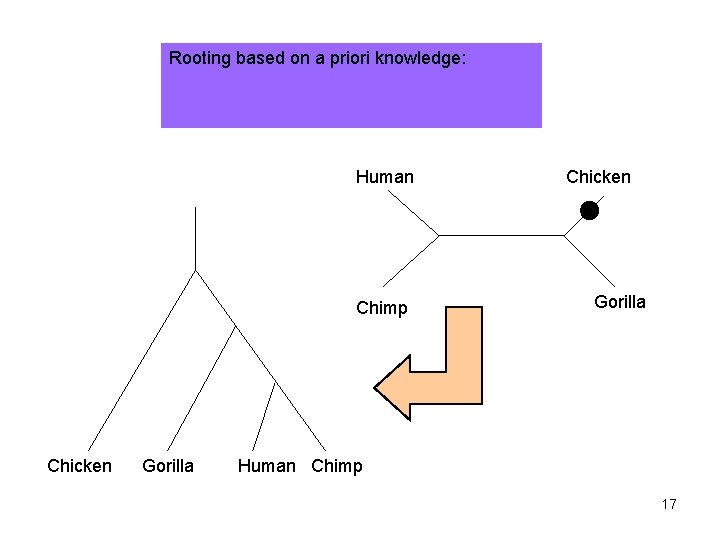
Rooting based on a priori knowledge: Human Chimp Chicken Gorilla Human Chimp 17

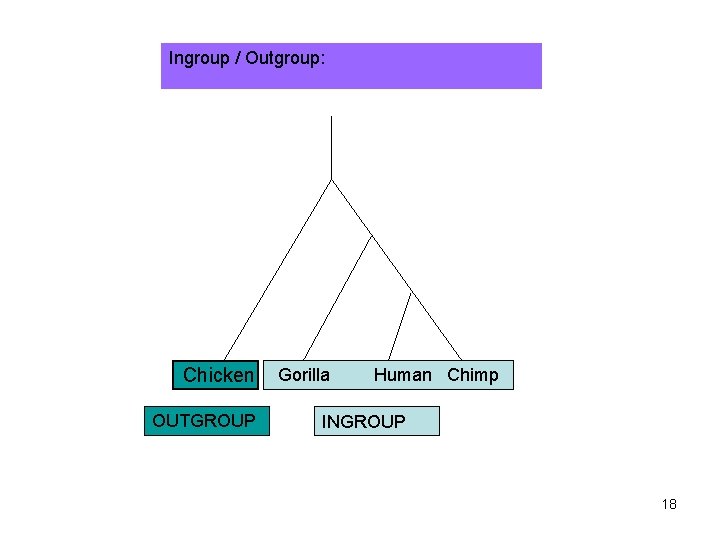
Ingroup / Outgroup: Chicken OUTGROUP Gorilla Human Chimp INGROUP 18

Tree of life

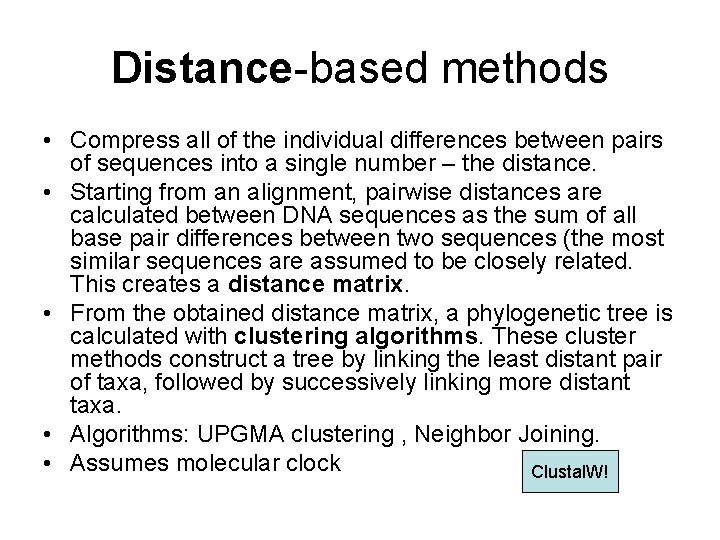
Distance-based methods • Compress all of the individual differences between pairs of sequences into a single number – the distance. • Starting from an alignment, pairwise distances are calculated between DNA sequences as the sum of all base pair differences between two sequences (the most similar sequences are assumed to be closely related. This creates a distance matrix. • From the obtained distance matrix, a phylogenetic tree is calculated with clustering algorithms. These cluster methods construct a tree by linking the least distant pair of taxa, followed by successively linking more distant taxa. • Algorithms: UPGMA clustering , Neighbor Joining. • Assumes molecular clock Clustal. W!

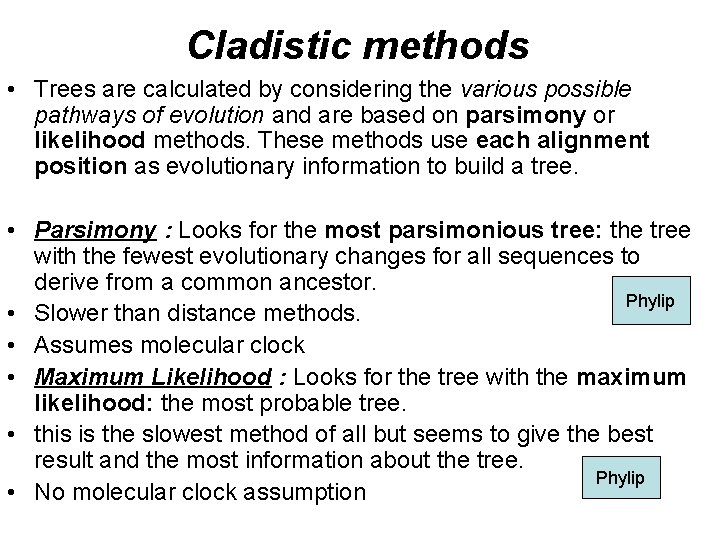
Cladistic methods • Trees are calculated by considering the various possible pathways of evolution and are based on parsimony or likelihood methods. These methods use each alignment position as evolutionary information to build a tree. • Parsimony : Looks for the most parsimonious tree: the tree with the fewest evolutionary changes for all sequences to derive from a common ancestor. Phylip • Slower than distance methods. • Assumes molecular clock • Maximum Likelihood : Looks for the tree with the maximum likelihood: the most probable tree. • this is the slowest method of all but seems to give the best result and the most information about the tree. Phylip • No molecular clock assumption

Even the best evolutionary models can't solve this problem. . . Two homologous DNA sequences which descended from an ancestral sequence and accumulated mutations since their divergence from each other. Note that although 12 mutations have accumulated, differences can be detected at only three nucleotide sites.

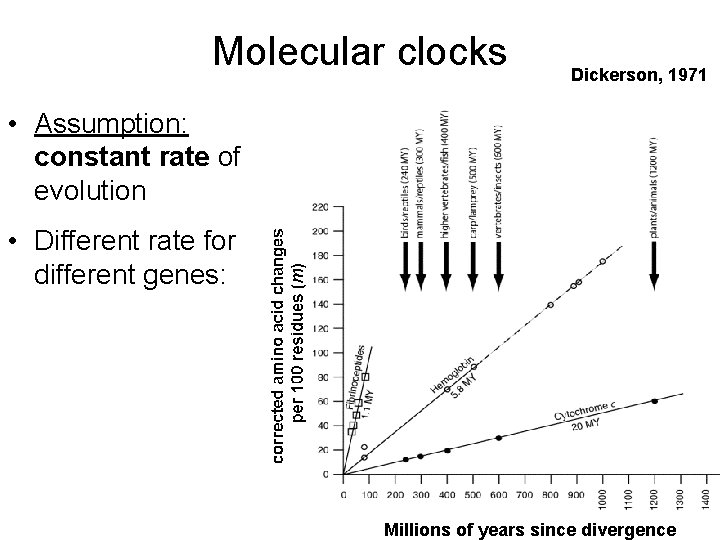
Molecular clocks Dickerson, 1971 • Assumption: constant rate of evolution • Different rate for different genes: Millions of years since divergence

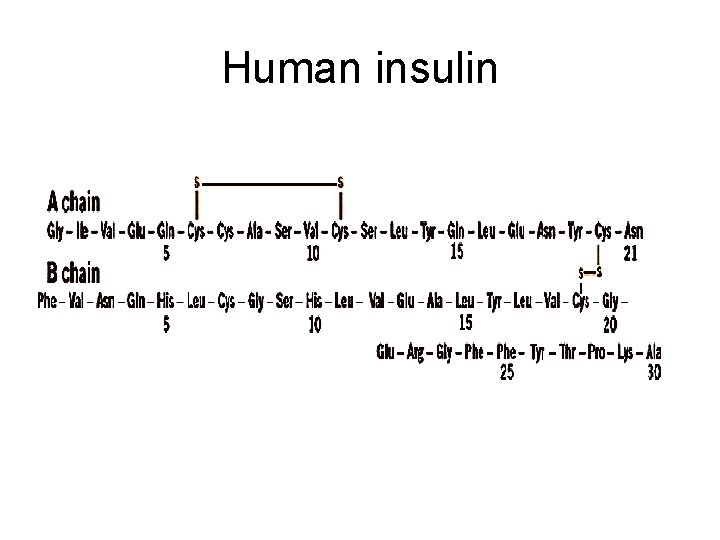
Human insulin

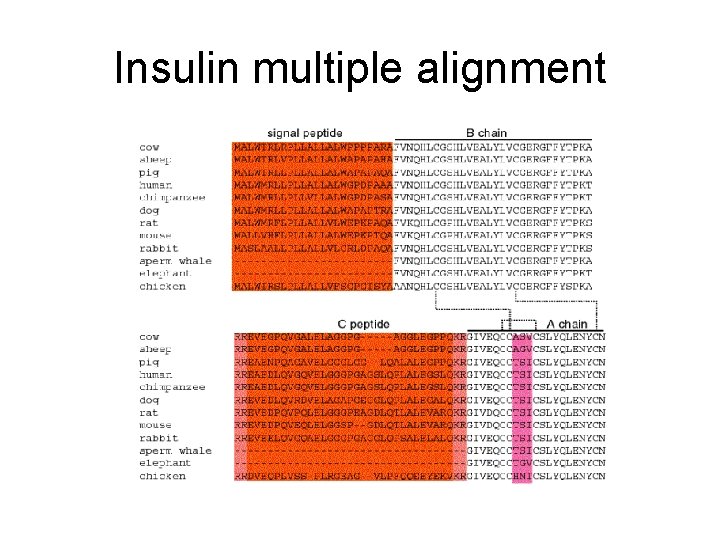
Insulin multiple alignment

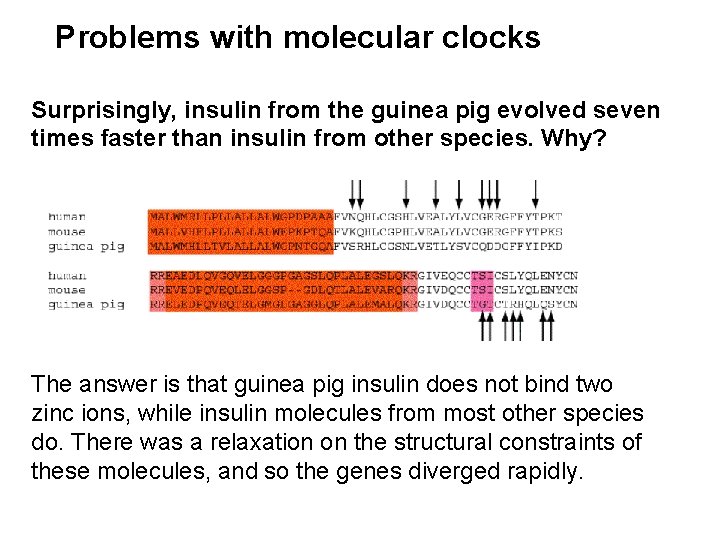
Problems with molecular clocks Surprisingly, insulin from the guinea pig evolved seven times faster than insulin from other species. Why? The answer is that guinea pig insulin does not bind two zinc ions, while insulin molecules from most other species do. There was a relaxation on the structural constraints of these molecules, and so the genes diverged rapidly.

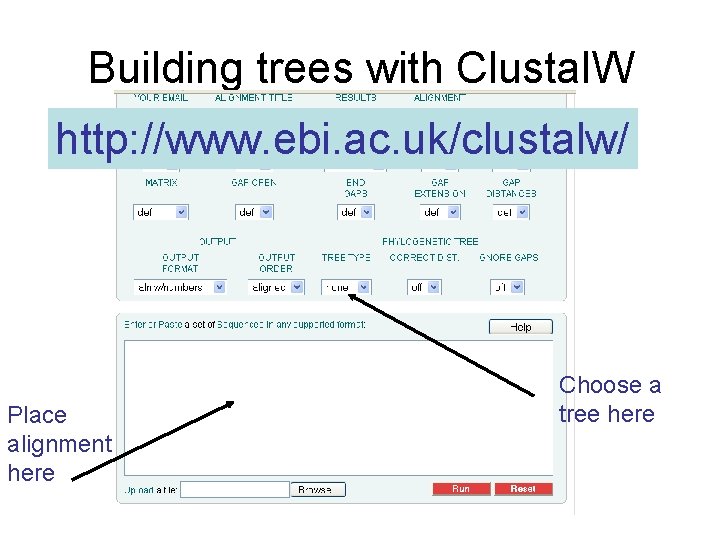
Building trees with Clustal. W http: //www. ebi. ac. uk/clustalw/ Place alignment here Choose a tree here

PHYLIP • A suite of phylogeny tools • Both web servers and stand-alone applications • Used for distance/parsimony/maximum likelihood • http: //bioweb. pasteur. fr/seqanal/phylogeny /phylip-uk. html

Sequences

Bootstrapping • Assigns confidence to individual tree branches • Columns of the alignment are randomly sampled (with replacement) and the tree is recomputed X many interactions • Boorstrap value of a branch = how many iterations had it.

Collections of homologous genes • Homologene @ Entrez – http: //www. ncbi. nlm. nih. gov/sites/entrez? db=homolog ene • COG – Clusters of Orthologous Genes – Results of Blast All-vs-All between genomes. Genes within the same COG are “pairwise best hits” – http: //www. ncbi. nlm. nih. gov/COG/ • RDP – Ribosomal sequences – The “standard” sequences for doing species phylogeny – Focused on Bacteria – http: //rdp 8. cme. msu. edu/html/

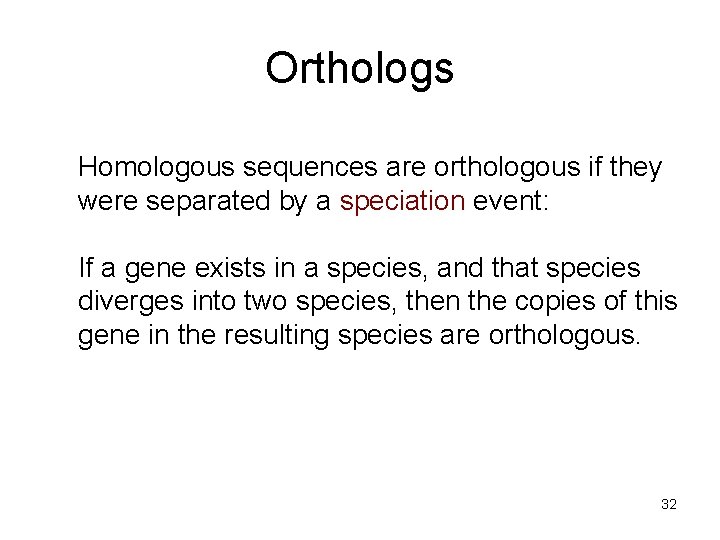
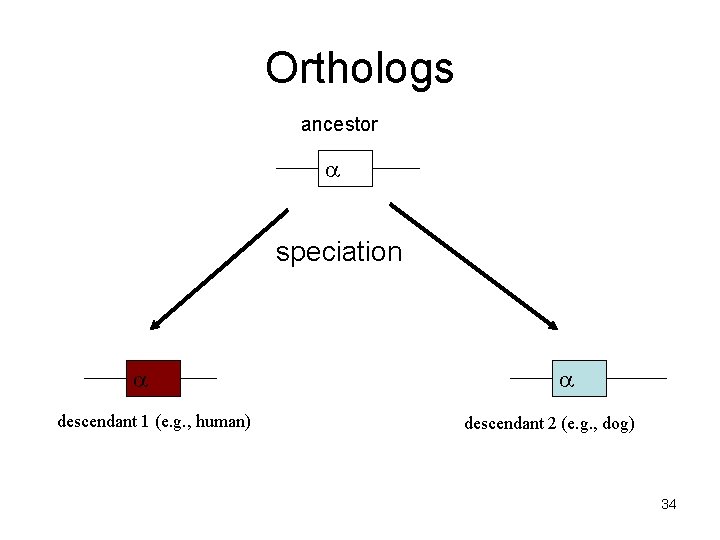
Orthologs Homologous sequences are orthologous if they were separated by a speciation event: If a gene exists in a species, and that species diverges into two species, then the copies of this gene in the resulting species are orthologous. 32

Orthologs • Orthologs will typically have the same or similar function in the course of evolution. • Identification of orthologs is critical for reliable prediction of gene function in newly sequenced genomes. 33

Orthologs ancestor a speciation a descendant 1 (e. g. , human) a descendant 2 (e. g. , dog) 34

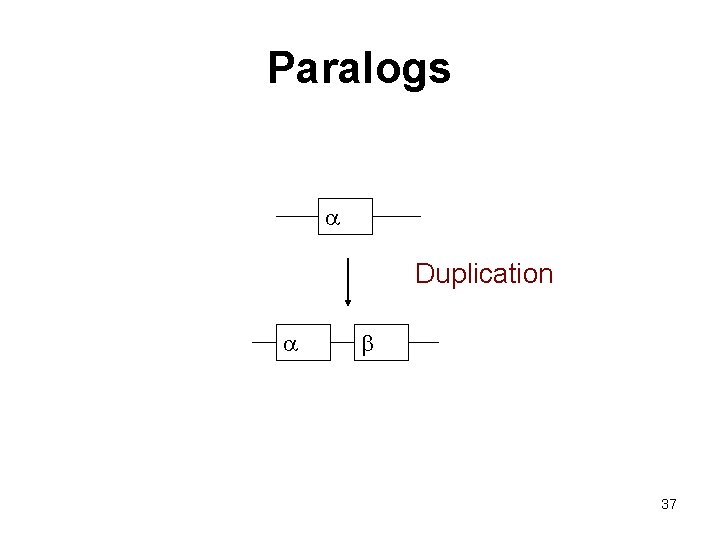
Paralogs Homologous sequences are paralogous if they were separated by a gene duplication event: If a gene in an organism is duplicated, then the two copies are paralogous. 35

Paralogs • Orthologs will typically have the same or similar function. • This is not always true for paralogs due to lack of the original selective pressure upon one copy of the duplicated gene, this copy is free to mutate and acquire new functions. 36

Paralogs a Duplication a b 37

(taken from NCBI) 38

Using BLAST and phylogeny to study gene evolution 39

Mol. Biol. Evol. (2005) 22: 598 -606 40

Evolutionary rate and conservation Functionally or structurally important sites are conserved: Conserved sites Variable sites “slow” evolving sites “fast evolving” sites Sites which are under a functional/structural constraint are conserved, and evolve slowly 41

Conservation in an MSA S 1 S 2 S 3 KITAYCELARTDMKLGLDFYKGVSLANWVCLAKWESGYN MPFERCELARTLKRMADADIRGVSLANWVCLAKWFWDGG MPFERCELARTLKRMMDADIRGVSLANWVCLAKWFWDGG From the MSA (and the tree), one can determine how conserved is a gene. 42

“Inverse relation between evolutionary rate and age of mammalian genes”: Protocol 43

Step 1 - BLAST Build the dataset of mammalian genes 44

Step 1 – BLAST: build the dataset of mammalian genes, based on mouse-human ortholog pairs • The orthologs are defined as pairs of reciprocal BLAST hits. • Eliminate genes with more than one potential orthologous sequence. • Select only genes which the human protein was functionally annotated. 45

Step 2 – Calculate conservation 46

Step 2 – Calculate Evolutionary Rates (Conservation) For each orthologous pair: • Alignment at the amino acid level. • Measure evolutionary rate The dataset contained 6, 776 human-mouse gene pairs. 47

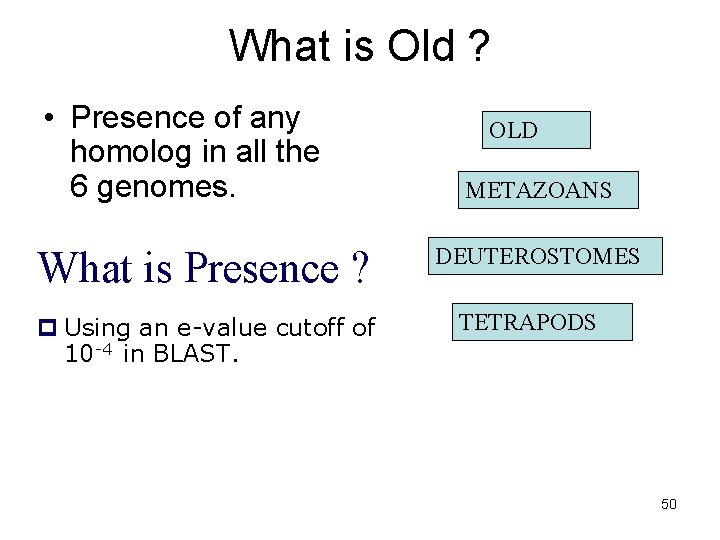
Step 3 – Assignment of Temporal Categories How old is each gene? Used BLAST to find homologs in 6 different eukaryotic genomes 48

Caenorhabditis elegans Drosophila melanogaster Takifugu rubripes Schizosaccharomyce s pombe Arabidopsis thaliana Saccharomyces cerevisiae 49

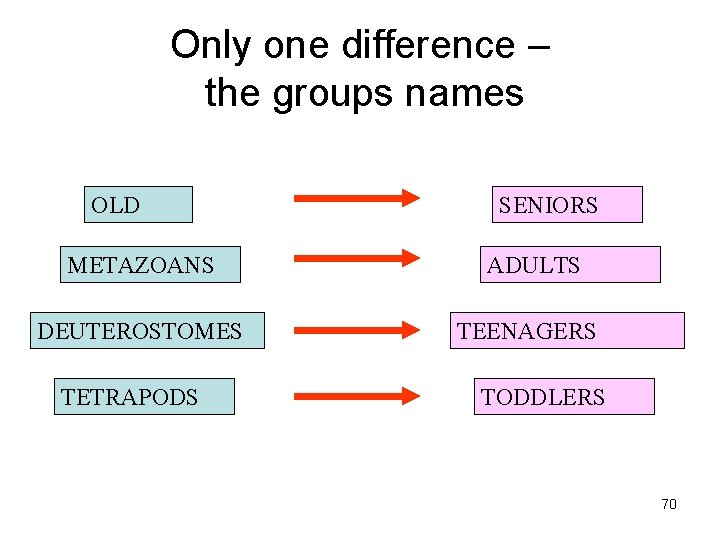
What is Old ? • Presence of any homolog in all the 6 genomes. What is Presence ? p Using an e-value cutoff of 10 -4 in BLAST. OLD METAZOANS DEUTEROSTOMES TETRAPODS 50

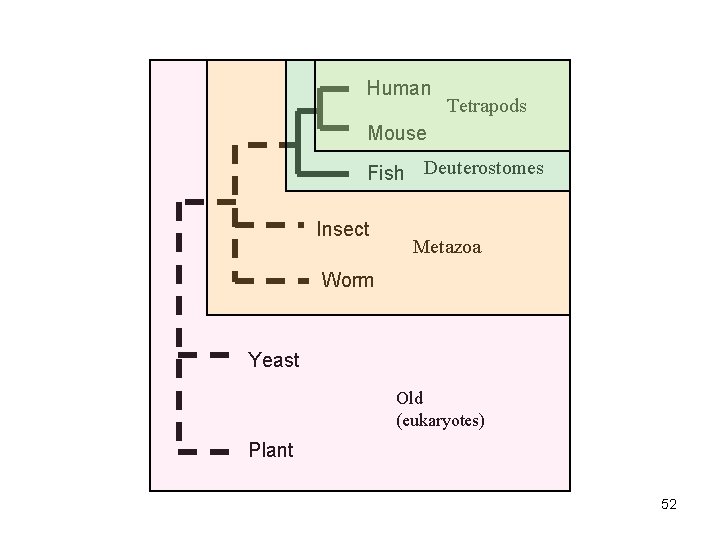
• METAZOANS - Organisms whose bodies consist of many cells, as distinct from Protozoa, which are unicellular; also commonly called animals. • DEUTEROSTOMES - The second of the two main groups of bilaterally symmetrical animals. The name derives from 'deutero' (second) 'stome' (mouth), referring to the origin of the definitive mouth as an opening independent from the blastopore of the embryo. • TETRAPODS - Any four-legged animals, including mammals, birds, reptiles and amphibians. 51

Human Tetrapods Mouse Fish Deuterostomes Insect Metazoa Worm Yeast Old (eukaryotes) Plant 52

Results 53

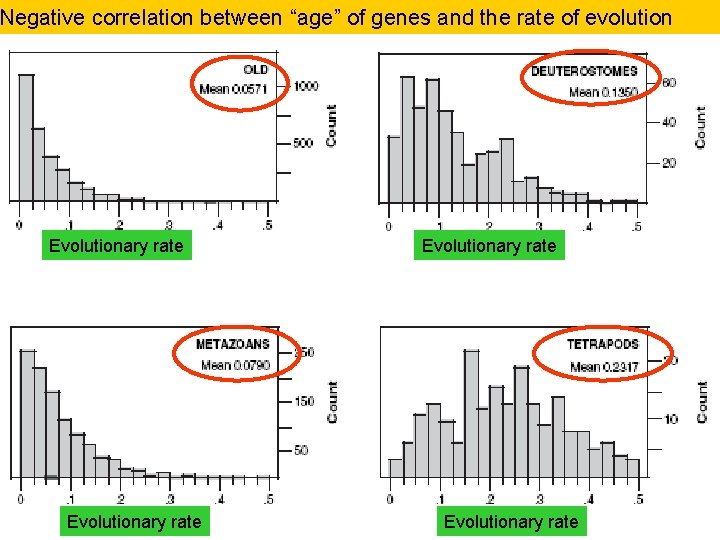
Negative correlation between “age” of genes and the rate of evolution Evolutionary rate 54

Control. • Changing the sensitivity of the BLAST detection to a more conservative one of 10 -10, did not significantly affect the result. 55

Explanations 56

• Functional constraints remained constant throughout the evolutionary history of each gene, but the newer genes are less constrained than older genes. • Functional constraints are not constant, rather they are weak at the time of origin of a gene and they become progressively more stringent with age. 57

Eran Elhaik, Niv Sabath, and Dan Graur 58 Mol. Biol. Evol. 23(1): 1– 3. 2006

Goal • To show that these results are an artifact caused by our inability to detect similarity when genetic distances are large. 59

Simulation 60

The evolutionary process Ala Arg … Val Ala Arg … Replacement probabilities Val Rat Mouse Cat Dog 61 Fly

The evolutionary process Ala Arg … Val Ala Arg … Replacement probabilities Val Rat Mouse Cat V Dog 62 Fly

The evolutionary process Ala Arg … Val Ala Arg … Replacement probabilities Val Rat V Mouse Cat V Dog 63 Fly

The evolutionary process Ala Arg … Val Ala Arg … Replacement probabilities Val Rat L V Mouse Cat V Dog 64 Fly

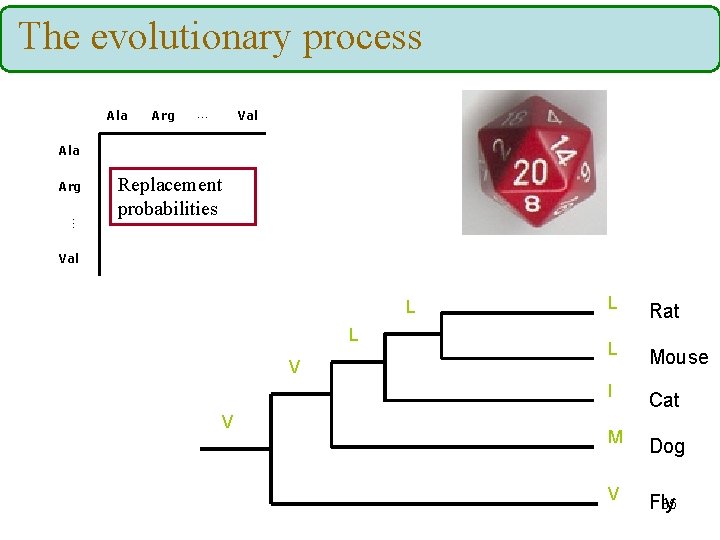
The evolutionary process Ala Arg … Val Ala Arg … Replacement probabilities Val L L V V L Rat L Mouse I Cat M Dog V 65 Fly

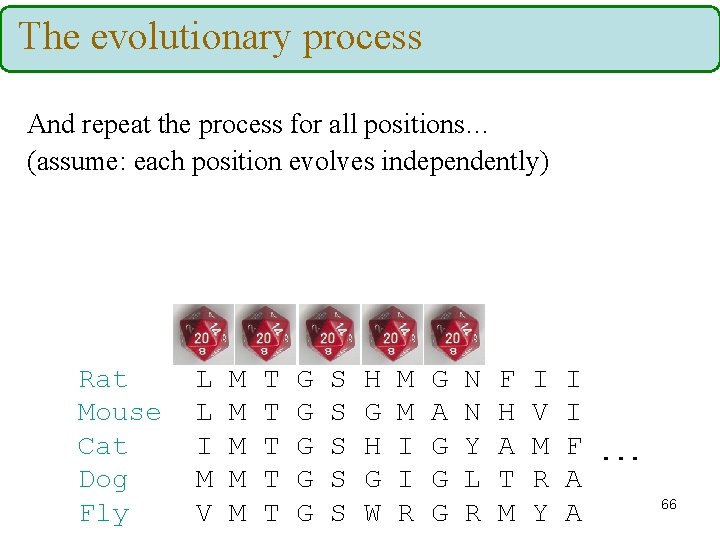
The evolutionary process And repeat the process for all positions… (assume: each position evolves independently) Rat Mouse Cat Dog Fly L L I M V M M M T T T G G G S S S H G W M M I I R G A G G G N N Y L R F H A T M I V M R Y I I F. . . A A 66

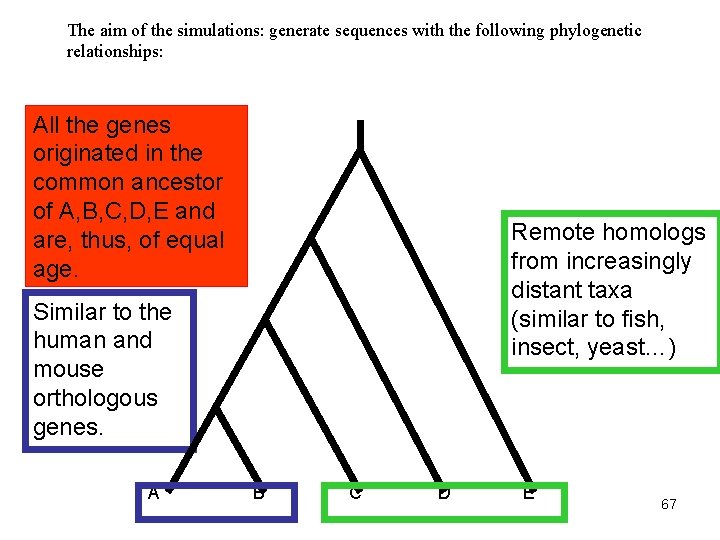
The aim of the simulations: generate sequences with the following phylogenetic relationships: All the genes originated in the common ancestor of A, B, C, D, E and are, thus, of equal age. Remote homologs from increasingly distant taxa (similar to fish, insect, yeast…) Similar to the human and mouse orthologous genes. A B C D E 67

Simulation • They simulated genes with 101 different rates. • High rate higher likelihood for an amino acid replacement in each branch. 68

After simulating the sequences: Use BLAST, at the same way that Alba and Castresana used it, to detect homology between gene A to genes C, D and E. 69

Only one difference – the groups names OLD METAZOANS DEUTEROSTOMES TETRAPODS SENIORS ADULTS TEENAGERS TODDLERS 70

Results 71

Same as Alba and Castresana 72

But all the simulated genes are at the same “age”. What is the problem ? ? ? 73

We can only count genes that are identified as homologous by the protocol … BLAST 74

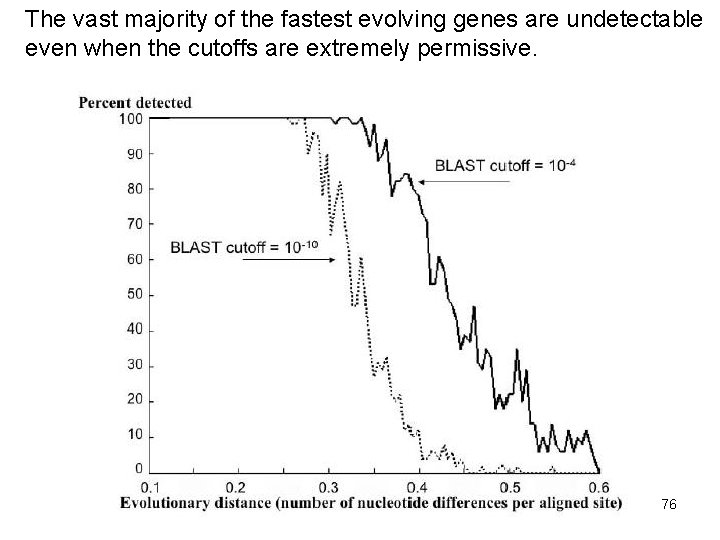
Alba and Castresana may have, thus, failed to spot the vast majority of homologs from among the fastest evolving genes 75

The vast majority of the fastest evolving genes are undetectable even when the cutoffs are extremely permissive. 76

Conclusion 77

The inverse relationship between evolutionary rate and gene age is an artifact caused by our inability to detect similarity when genetic distances are large. 78

• Since genetic distance increases with time of divergence and rate of evolution, it is difficult to identify homologs of fast evolving genes in distantly related taxa. • Thus, fast evolving genes may be misclassified as “new”. 79

So, the only conclusion Slowly evolving that can be drawn from genes Alba and Castresana’s study is that evolve slowly !!! 80