Phylo Pat phylogenetic pattern analysis of eukaryotic genes











- Slides: 22

Phylo. Pat phylogenetic pattern analysis of eukaryotic genes Tim Hulsen 2006 -11 -22

Goal • Create database of sequence (in this case: gene, Protein World: protein) relationships over several species • Can be used for transferring information from model organisms to humans (-> drug testing) • Database can be used for many other things too… like: analysis using phylogenetic patterns

Introduction (1) • Phylogenetic patterns show presence/absence of genes over a certain set of species: e. g. for 10 species: 0011101011 • Very useful for all kinds of evolutionary analyses: – Origin of certain genes – Deletion of certain genes – Clustering of genes with similar patterns: likely to have similar function / be in same pathway

Introduction (2) • Earlier phylogenetic pattern initiatives: – Phylogenetic Pattern Search (PPS), incorporated into COG (Natale et al. , 2000) – Extended Phylogenetic Patterns Search (EPPS) (Reichard & Kaufmann, 2003) – Incorporated into Ortho. MCL-DB (Chen et al. , 2006) • All applied on proteins, not on genes! • Phylo. Pat: phylogenetic pattern analysis of eukaryotic genes

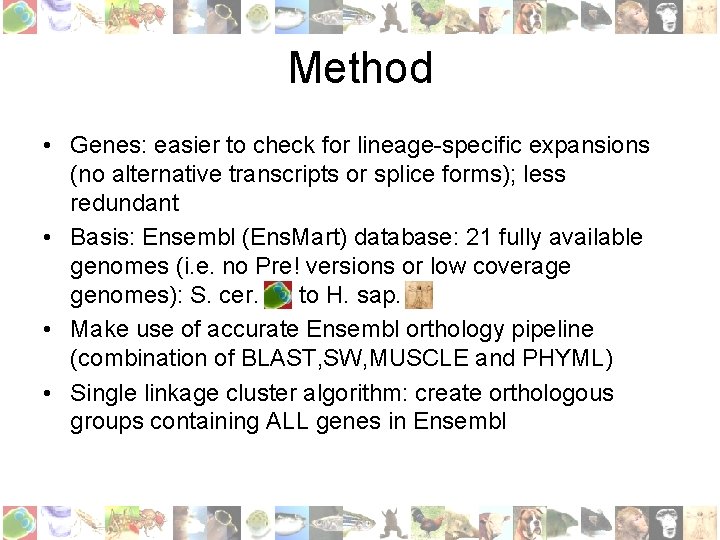
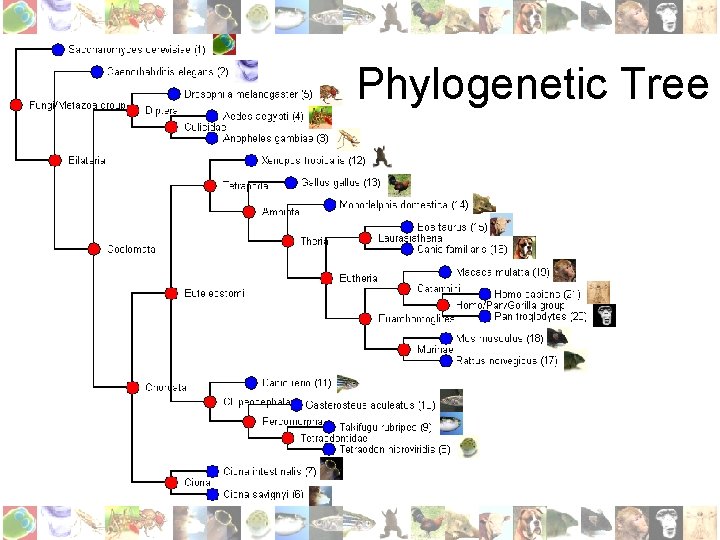
Method • Genes: easier to check for lineage-specific expansions (no alternative transcripts or splice forms); less redundant • Basis: Ensembl (Ens. Mart) database: 21 fully available genomes (i. e. no Pre! versions or low coverage genomes): S. cer. to H. sap. • Make use of accurate Ensembl orthology pipeline (combination of BLAST, SW, MUSCLE and PHYML) • Single linkage cluster algorithm: create orthologous groups containing ALL genes in Ensembl

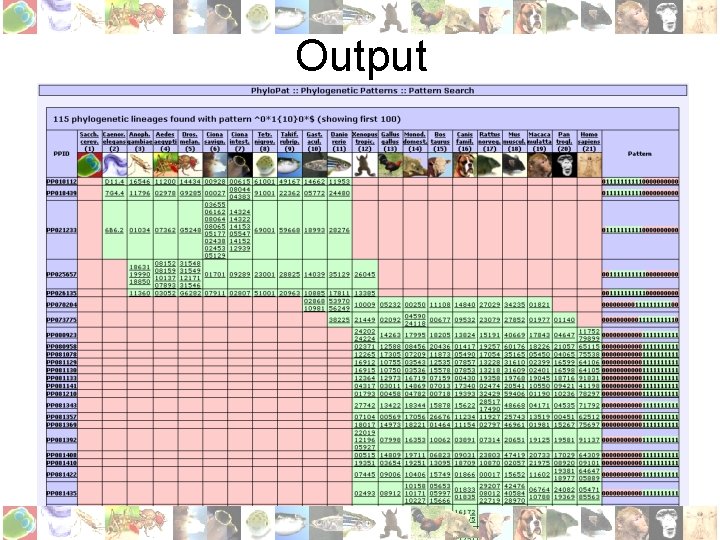
Results • 446, 825 genes were clustered into 147, 922 groups, using 3, 164, 088 orthologies from 21 species • Species ordered from ‘low’ ( ) to ‘high’ ( ), i. e. approximate distance to human : • Can be queried in several ways • Output in HTML, Excel or plain text format

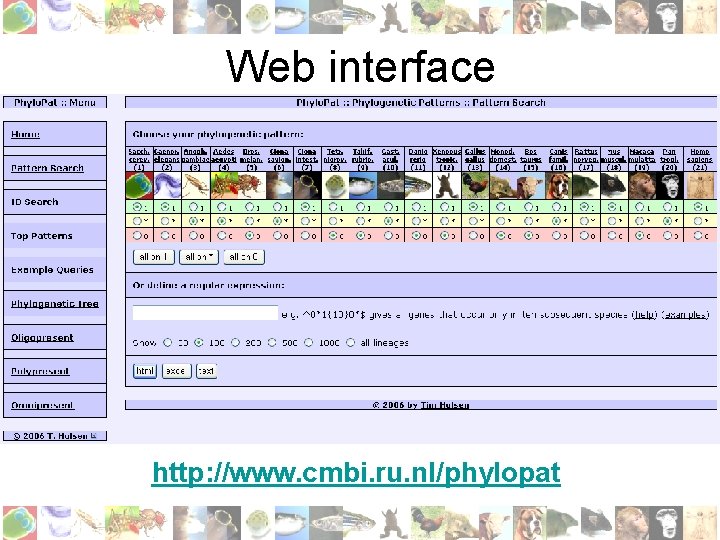
Web interface http: //www. cmbi. ru. nl/phylopat

Pattern/ID Search • Binary string: 0=absent, 1=present, *=absent/present e. g. ‘ 00000****1111’: must be absent in non-chordata , must be present in all mammals • My. SQL regular expression: e. g. ‘^0*1{10}0*$’ gives all genes that occur only in ten subsequent species • Input list of Ensembl/EMBL IDs (Phylo. Pat contains EMBL to Ensembl mapping)

Output

Phylogenetic Tree

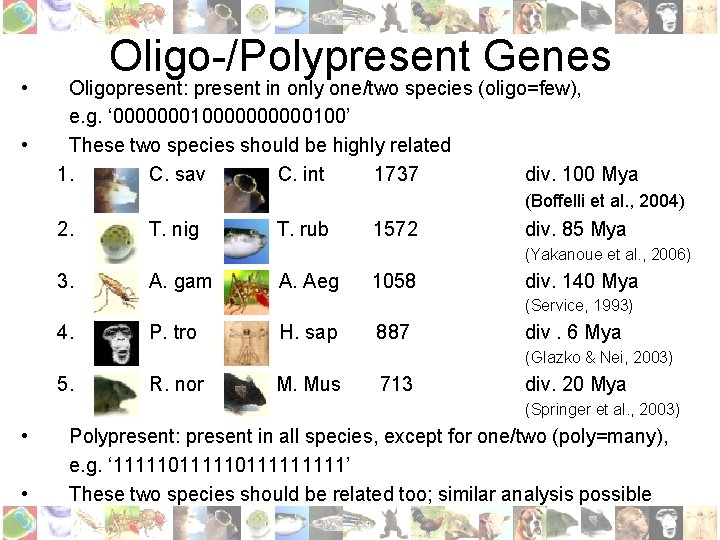
• • Oligo-/Polypresent Genes Oligopresent: present in only one/two species (oligo=few), e. g. ‘ 0000000100000100’ These two species should be highly related 1. C. sav C. int 1737 div. 100 Mya (Boffelli et al. , 2004) 2. T. nig T. rub 1572 div. 85 Mya (Yakanoue et al. , 2006) 3. A. gam A. Aeg 1058 div. 140 Mya (Service, 1993) 4. P. tro H. sap 887 div. 6 Mya (Glazko & Nei, 2003) 5. R. nor M. Mus 713 div. 20 Mya (Springer et al. , 2003) • • Polypresent: present in all species, except for one/two (poly=many), e. g. ‘ 11111011111’ These two species should be related too; similar analysis possible

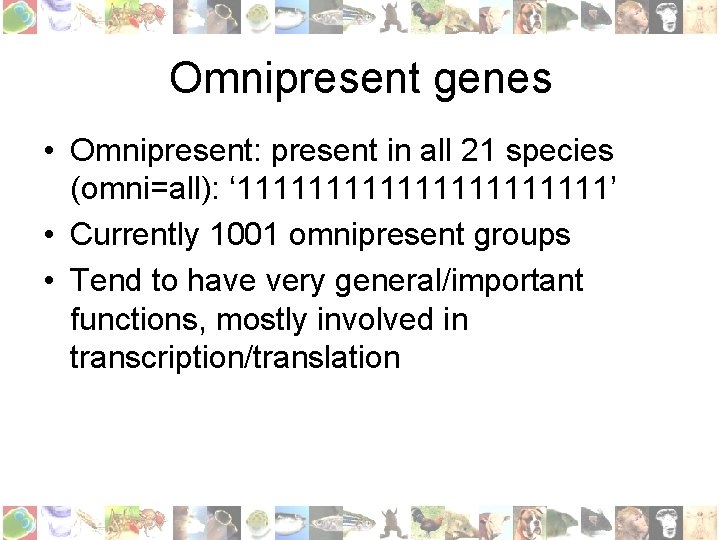
Omnipresent genes • Omnipresent: present in all 21 species (omni=all): ‘ 11111111111’ • Currently 1001 omnipresent groups • Tend to have very general/important functions, mostly involved in transcription/translation

Fati. GO analysis • Fati. GO: connection with GO terms, KEGG pathways, Inter. Pro domains, etc. (El-Shahrour et al. , 2004) • Analysis of all human genes in output by just one mouse click • e. g. omnipresent genes:

Other possibilities • Anti-correlating patterns: e. g. ‘ 0011111000000000’ and ‘ 1100000111111111’ could be completely different, or very similar (analogous)! • Easy homology-inferred functional annotation (using information from other genes in the same lineage)

Case study: Hox genes (1) • Hox genes determine where limbs and other body segments will grow in a developing embryo • Should exist mostly in vertebrates • Expansion in teleost fish species ( , 8 -11); seven Hox clusters instead of the mammalian four • Search Ensembl database for human genes with term ‘hox’ in annotation • 44 genes found -> enter in Phylo. Pat -> 32 groups found (PP######)

Case study: Hox genes (2) PPID PP 022041 PP 024984 PP 027791 PP 049478 PP 053824 PP 053827 PP 053828 PP 053829 PP 053830 PP 053832 PP 053833 PP 053834 PP 053835 PP 053836 PP 053838 PP 053839 PP 053840 PP 053842 PP 053844 PP 053845 PP 053846 PP 053847 PP 053849 PP 053853 PP 053854 PP 053858 PP 070659 PP 075622 PP 084287 PP 085049 PP 087941 PP 089685 # genes per species 011111136562233233222 00100001111111 00111002334323333 000000221153112322223 00000001112001011 0000000222111111 000000021111212122222 00000006334112222 000000011110010111111 00000002111101111 000000021110111111011 00000003110101111 000000021110111111101 000000021111111 000000021110101111111 0000000111110111 000000021111201011101 0000000432211111 00000003223101111 0000000211111011 0000000211211111 0000000222111111 000000034151132333323 00000001111111011 000000032252223133213 00000001112001111 00000121212222222 0000010001111111 0000001101111111 0000001011011111 00000000000011111 phylogenetic pattern 01111111111 0010000111111111 00000011111111 00000001111001011 000000011111111111111 00000001111111 000000011110010111111 000000011111111 0000000111111011 00000001110101111 0000000111111101 00000001111111 000000011110101111111 0000000111110111 000000011111101011101 000000011111111 0000000111111011 000000011111111111111 00000001111111011 000000011111111 00000111111 0000010001111111 0000001101111111 0000001011011111 00000000000011111 gene name(s) MSX 1, MSX 2 HOXC 4 TLX 1, TLX 2, TLX 3 HOXB 8, HOXC 8, HOXD 8 HOXD 11 HOXA 10 HOXC 13, HOXD 13 HOXA 1, HOXB 1 HOXB 4 HOXA 5 HOXB 2 HOXD 3 HOXA 9 HOXA 3 HOXC 12 HOXD 4 HOXC 11 HOXA 13 HOXB 5 HOXB 3 HOXD 10 HOXA 2 HOXA 6, HOXB 6, HOXC 6 HOXA 4 HOXB 9, HOXC 9, HOXD 9 HOXA 11 HOXA 7, HOXB 7 HOXC 5 HOXC 10 HOXD 12 HOXB 13

Case study: Hox genes (3) PPID(s) PP 053829, 085049 PP 053847, 053833 PP 053836, 053845, 053834 PP 053832, 053844, 075622 PP 053849 PP 053835, 053854 PP 053827, 084287, 053846 PP 053858, 053840, 053824 PP 053838, 087941 PP 053842, 089685, 053828 name HOX 1 HOX 2 HOX 3 HOX 5 HOX 6 HOX 9 HOX 10 HOX 11 HOX 12 HOX 13 cl. A HOXA 1 HOXA 2 HOXA 3 HOXA 5 HOXA 6 HOXA 9 HOXA 10 HOXA 11 cl. B HOXB 1 HOXB 2 HOXB 3 HOXB 5 HOXB 6 HOXB 9 cl. C PP 053853, 053830, 024984, 053839 HOX 4 PP 027791 TLX HOXA 4 TLX 1 HOXB 4 TLX 2 HOXC 4 TLX 3 PP 070659 HOX 7 HOXA 7 HOXB 7 PP 049478 HOX 8 PP 022041 MSX HOXD 3 HOXC 5 HOXC 6 HOXC 9 HOXC 10 HOXC 11 HOXC 12 HOXA 13 HOXB 13 HOXC 13 HOXB 8 MSX 1 MSX 2 cl. D HOXD 1 HOXC 8 HOXD 9 HOXD 10 HOXD 11 HOXD 12 HOXD 13 HOXD 4 HOXD 8 first sp. T. nigrov. position anterior PG 3 central posterior posterior A. gamb. central G. acul. central Vertebrate C. intest. central Nonvertebrate C. eleg. Nonvertebrate ‘First’ vertebrate Nonvertebrate

Conclusions • Phylo. Pat: quick and easy tool for phylogenetic pattern search on complete Ensembl database • Also usable for study of lineage-specific expansions of genes • Just updated to Ensembl v 41 (released last Thursday); 5 new species: D. nov E. tel L. afr O. cun O. lat + extra option: gene neighborhood

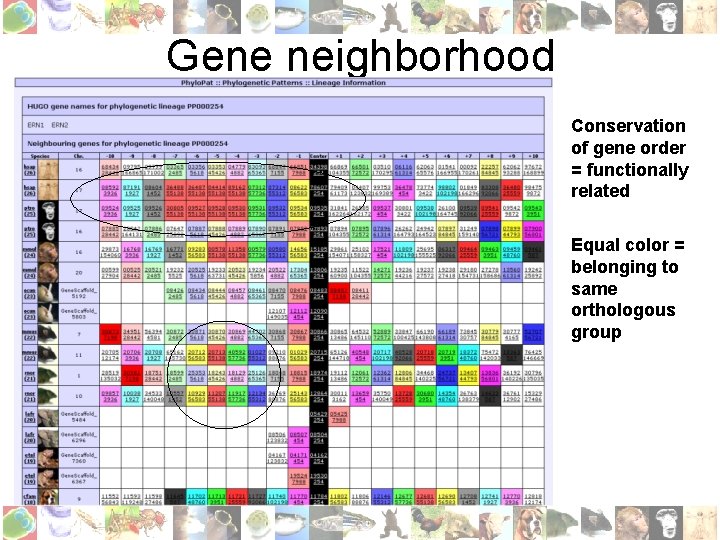
Gene neighborhood Conservation of gene order = functionally related Equal color = belonging to same orthologous group

Future directions • Map (drug discovery) pathways in model organisms and man to each other, to understand differences between species • Now being applied in immunogenomics study within Organon: how does immune system evolve from model organisms to man?

Acknowledgements Supervision: • Peter Groenen • Jacob de Vlieg Fruitful discussions: • Wilco Fleuren • Erik Franck • Nanning de Jong • Arnold Kuzniar supervisor head of group suggestions

Where to find • Web interface: http: //www. cmbi. ru. nl/phylopat (accessible through www. cmbi. ru. nl and www. nbic. nl) • Publication: Hulsen T. , Groenen P. M. A. , de Vlieg J. BMC Bioinformatics 2006, 7: 398 http: //www. biomedcentral. com/1471 -2105/7/398 • Powered by Ensembl: http: //www. ensembl. org/info/about/ensembl_powered. html