Phylgenetic tree 70 identity 85 identity 60 identity





- Slides: 11

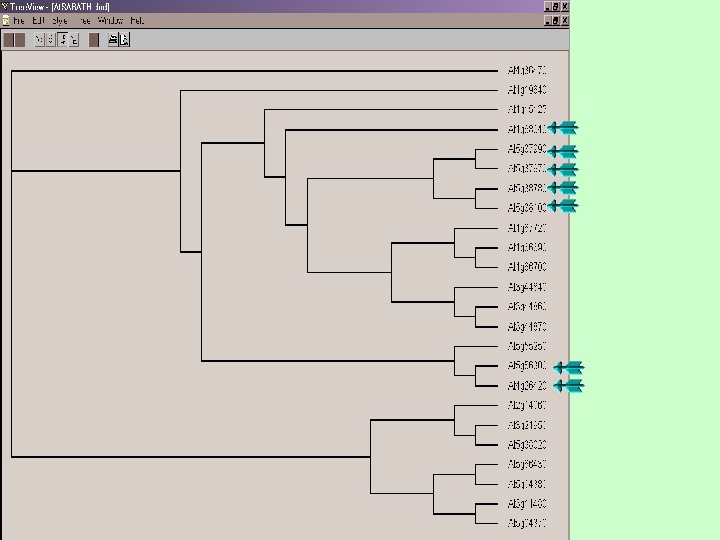
Phylgenetic tree

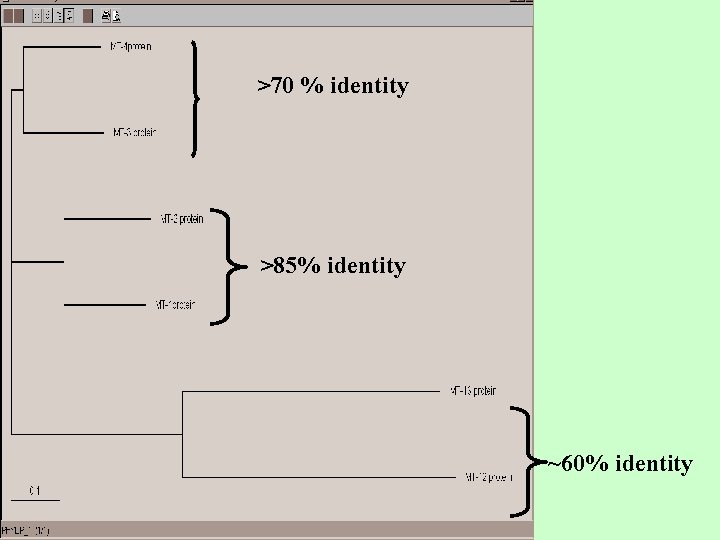
>70 % identity >85% identity ~60% identity

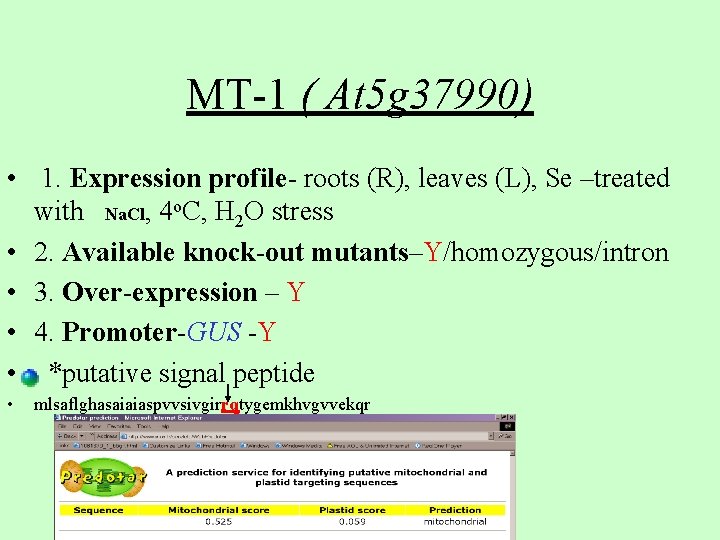
MT-1 ( At 5 g 37990) • 1. Expression profile- roots (R), leaves (L), Se –treated with Na. Cl, 4 o. C, H 2 O stress • 2. Available knock-out mutants–Y/homozygous/intron • 3. Over-expression – Y • 4. Promoter-GUS -Y • *putative signal peptide • mlsaflghasaiaiaspvvsivgirrqtygemkhvgvvekqr

MT-2 ( At 5 g 37970) • 1. Expression profile – roots (R), leaves (L), Se – treated with 4 o. C, H 2 O stress • 2. Knock-out mutants-NO • 3. Over-expression - YES • 4. Promoter-GUS - YES • *putative signal peptide • Mlsafwghasaiaiaspvvsivgfqgdrttirrptygemkfvdvverpr – no predicted cleveage signal

MT-3 ( At 5 g 38780) • 1. Expression profile- R, Leaves, Se. Na. Cl, Se 4 C, Water treated leaves • 2. Available knock-out mutants-Y-homozygous/exon • 3. Over-expression - Y • 4. Promoter-GUS – Y o • *Microarray: detected in senescent leaves /SA, JA mutants/

MT-4 ( At 5 g 38100) • • 1. Expression profile-St, L, F, R, Se-Na. Cl, Se-4 C 2. Available knock-out mutants-? -one putative, 5`UTR 3. Over-expression - Y 4. Promoter-GUS - Y o

MT-9 ( At 5 g 68040) • • 1. Expression profile - R 2. Available knock-out mutants-Y/homozygous/exon 3. Over-expression - Y 4. Promoter-GUS - Y

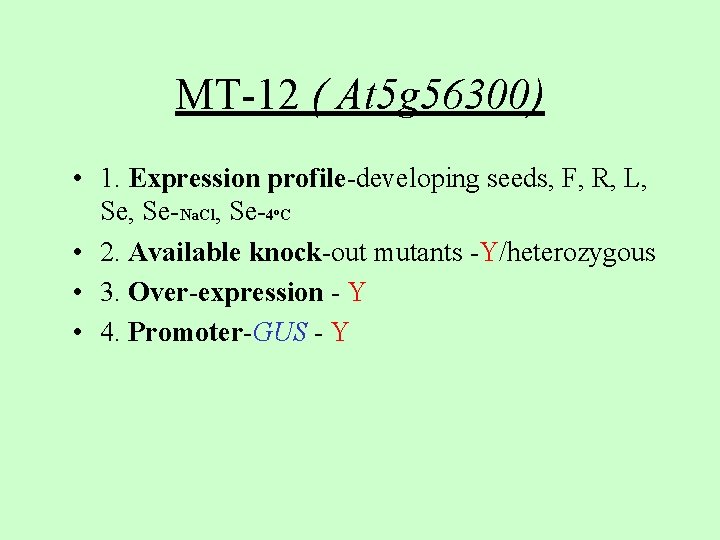
MT-12 ( At 5 g 56300) • 1. Expression profile-developing seeds, F, R, L, Se-Na. Cl, Se-4 C • 2. Available knock-out mutants -Y/heterozygous • 3. Over-expression - Y • 4. Promoter-GUS - Y o

MT-13 ( At 5 g 26420) • • 1. Expression profile-exclusively in developing seeds 2. Available knock-out mutants-? two putative mutants 3. Over-expression - Y 4. Promoter-GUS -Y

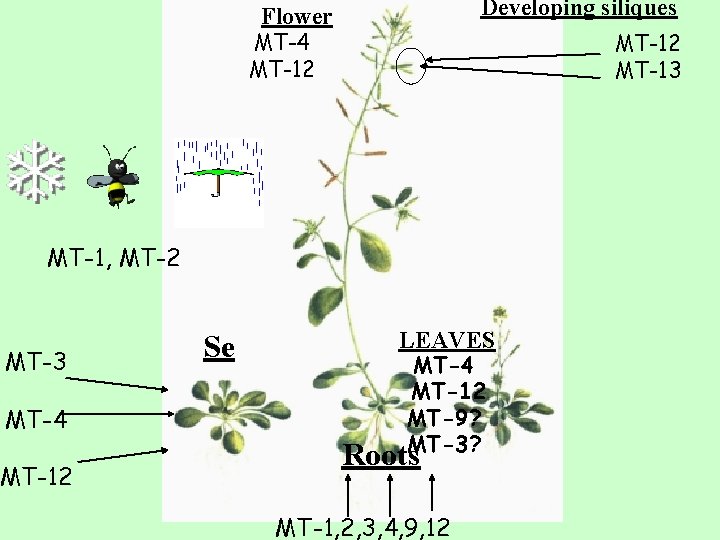
Developing siliques Flower MT-4 MT-12 Back to the first MT-1, MT-2 MT-3 MT-4 MT-12 Se LEAVES MT-4 MT-12 MT-9? MT-3? Roots MT-1, 2, 3, 4, 9, 12 MT-13

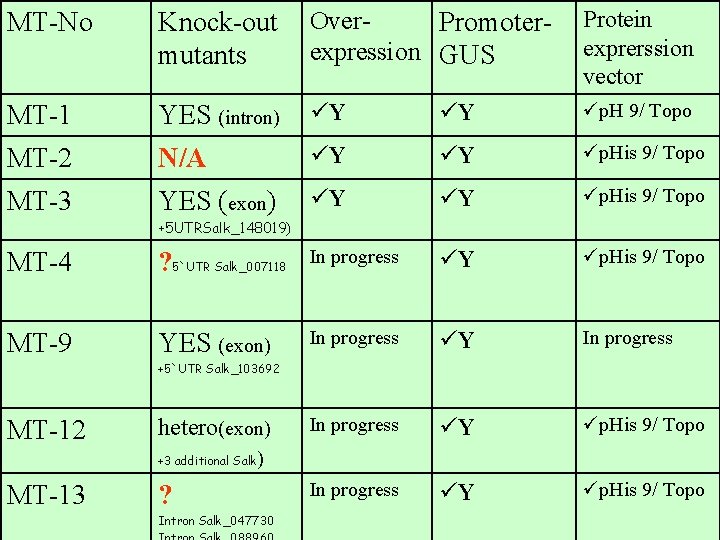
MT-No Knock-out mutants Over. Promoterexpression GUS Protein exprerssion vector MT-1 YES (intron) üY üY üp. H 9/ Topo MT-2 N/A üY üY üp. His 9/ Topo MT-3 YES (exon) üY üY üp. His 9/ Topo +5 UTRSalk_148019) MT-4 ? 5`UTR Salk_007118 In progress üY üp. His 9/ Topo MT-9 YES (exon) In progress üY üp. His 9/ Topo In progress üY üp. His 9/ Topo +5`UTR Salk_103692 MT-12 hetero(exon) +3 additional Salk MT-13 ) ? Intron Salk_047730
 Identity map
Identity map Bwtree
Bwtree Problem tree solution tree
Problem tree solution tree How about the orchid to a tree is a tree benefited
How about the orchid to a tree is a tree benefited Loser tree algorithm
Loser tree algorithm Species tree
Species tree Objective tree sample
Objective tree sample Difference between general tree and binary tree
Difference between general tree and binary tree Mckinsey problem solving approach
Mckinsey problem solving approach Convert 2-3-4 tree to red black
Convert 2-3-4 tree to red black Difference between plus tree and elite tree
Difference between plus tree and elite tree Problem tree and objective tree
Problem tree and objective tree





















