Phy CMAP Predicting protein contact map using evolutionary





- Slides: 36

Phy. CMAP: Predicting protein contact map using evolutionary and physical constraints by integer programming Zhiyong Wang and Jinbo Xu Toyota Technological Institute at Chicago Web server at http: //raptorx. uchicago. edu See http: //arxiv. org/abs/1308. 1975 for an extended version

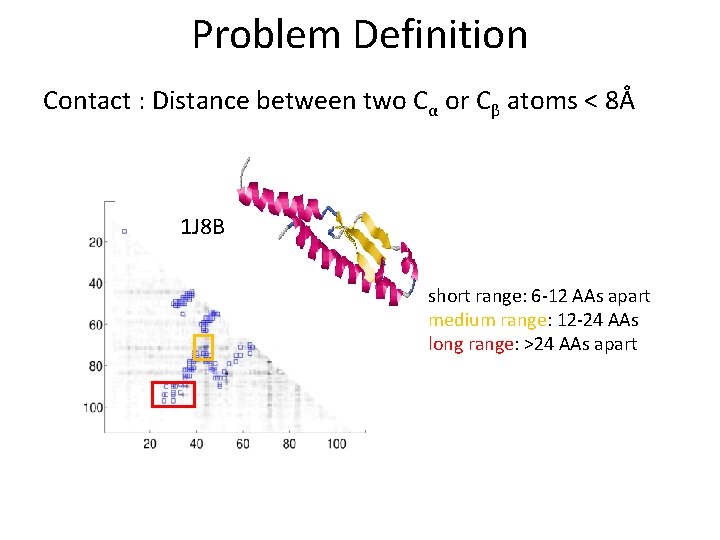
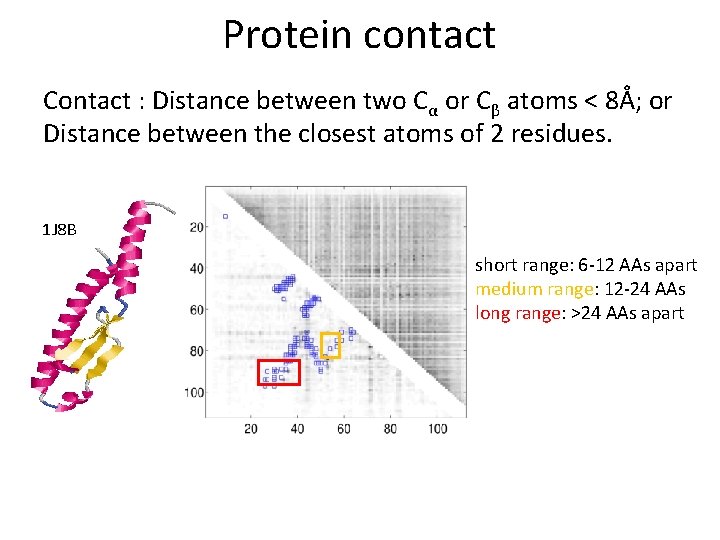
Problem Definition Contact : Distance between two Cα or Cβ atoms < 8Å 1 J 8 B short range: 6 -12 AAs apart medium range: 12 -24 AAs long range: >24 AAs apart

Existing Work Residue co-evolution method: mutual information (MI), PSICOV, Evfold ü Needs a large number of homologous sequences ü ü PSICOV and Evfold better than MI since they differentiate direct and indirect residue couplings (Residues A and C indirect coupling if it is due to direct A-B and B-C couplings) PSICOV and Evfold also enforce sparsity Supervised learning method: NNcon, SVMcon, CMAPpro ü Mutual information, sequence profile and others ü Predicts contacts one by one, ignoring their correlation ü Do not differentiate direct and indirect residue couplings First-principle method: Astro-Fold ü No evolutionary information ü Minimize contact potential ü Enforce physical feasibility including sparsity

Our Method: Phy. CMAP 1. Focus on proteins with few sequence homologs Ø proteins with many sequence homologs very likely have similar templates in PDB 2. Integrate by machine learning Ø seq profile, residue co-evolution and non-evolutionary info Ø (implicitly) differentiate direct and indirect residue couplings through feature engineering 3. Enforce physical constraints, which imply sparsity

Info used by Random Forests • Evolution info from a single protein family – sequence profile – co-evolution: 2 types of mutual information (MI) • Non-evolution info from the whole structure space: residue contact potential • Mixed info from the above 2 sources – homologous pairwise contact score – EPAD: context-specific evolutionary-based distancedependent statistical potential • amino acid physic-chemical properties

Mutual Information 1. Contrastive Mutual Information (CMI): remove local background by measuring the MI difference of one pair with its neighbors. 2. Chaining effect of residue couplings: MI, MI 2, MI 3, MI 4, equivalent to (1 -MI), (1 -MI)2, (1 -MI)3, (1 -MI)4 (see http: //arxiv. org/abs/1308. 1975 for more details)

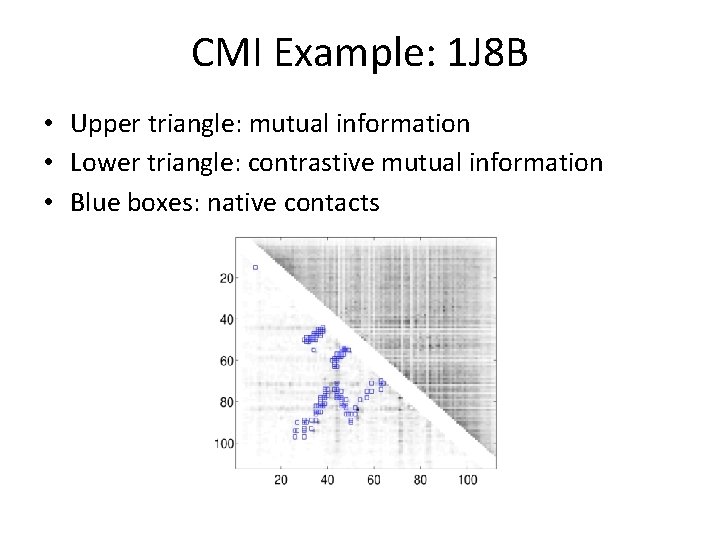
CMI Example: 1 J 8 B • Upper triangle: mutual information • Lower triangle: contrastive mutual information • Blue boxes: native contacts

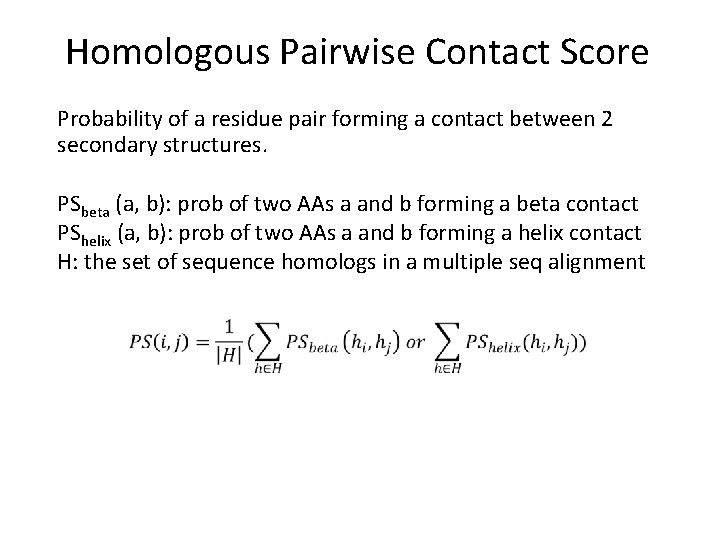
Homologous Pairwise Contact Score Probability of a residue pair forming a contact between 2 secondary structures. PSbeta (a, b): prob of two AAs a and b forming a beta contact PShelix (a, b): prob of two AAs a and b forming a helix contact H: the set of sequence homologs in a multiple seq alignment

Training Random Forests • Training dataset – – Chosen before CASP 10 started 900 non-redundant protein structures <25% sequence identity All contacts and 20% of non-contacts • Model parameters – Number of features: 300 – Number of trees: 500 – 5 fold cross validation

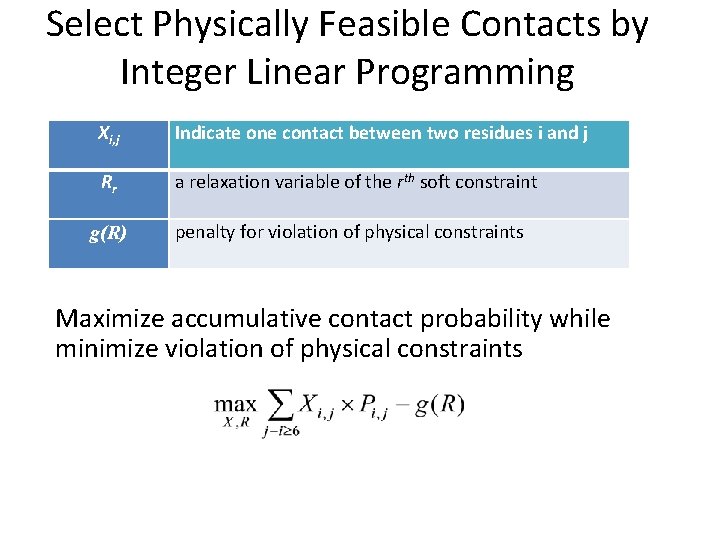
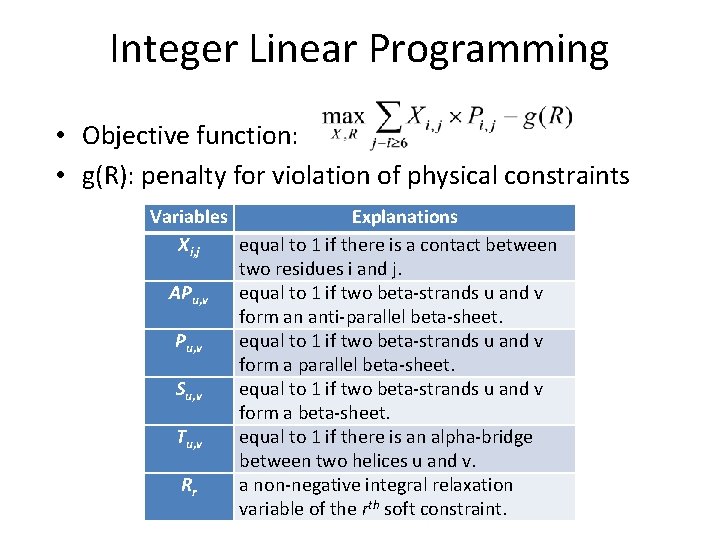
Select Physically Feasible Contacts by Integer Linear Programming Xi, j Indicate one contact between two residues i and j Rr a relaxation variable of the rth soft constraint g(R) penalty for violation of physical constraints Maximize accumulative contact probability while minimize violation of physical constraints

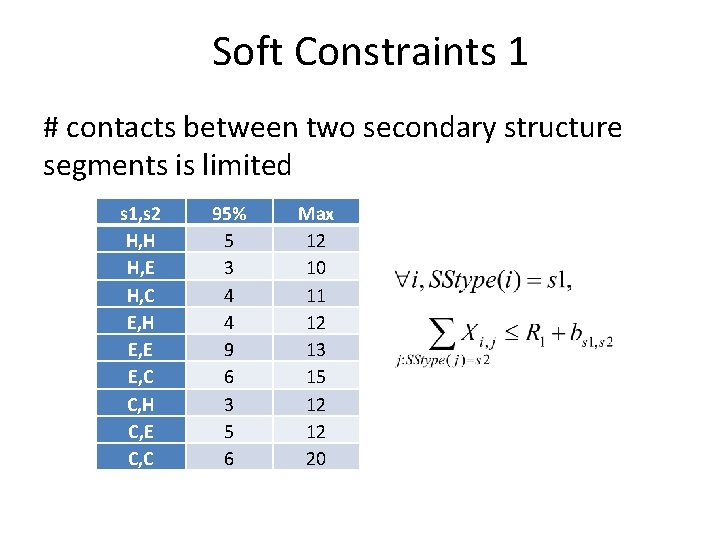
Soft Constraints 1 # contacts between two secondary structure segments is limited s 1, s 2 H, H H, E H, C E, H E, E E, C C, H C, E C, C 95% 5 3 4 4 9 6 3 5 6 Max 12 10 11 12 13 15 12 12 20

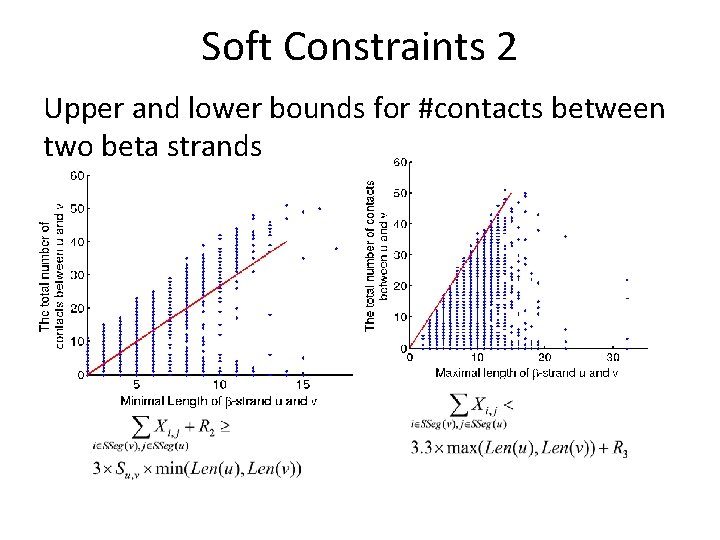
Soft Constraints 2 Upper and lower bounds for #contacts between two beta strands

Soft Constraints 3 Statistics shows that only 3. 4% of loop segments that have a contact between the start and end residues.

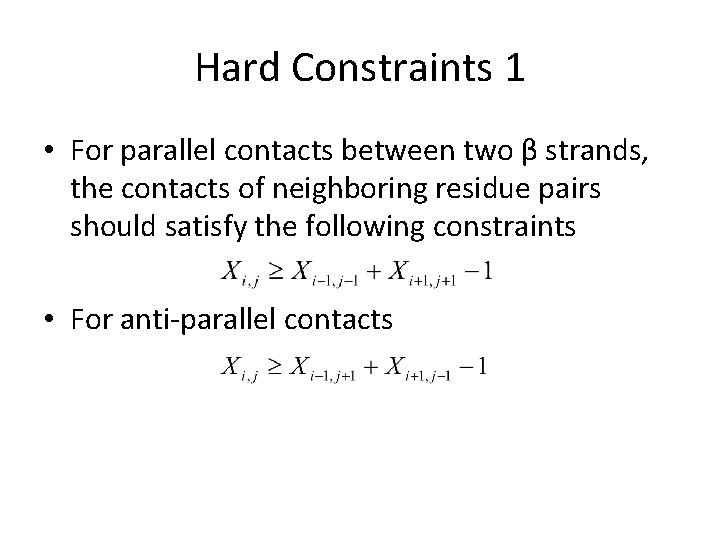
Hard Constraints 1 • For parallel contacts between two β strands, the contacts of neighboring residue pairs should satisfy the following constraints • For anti-parallel contacts

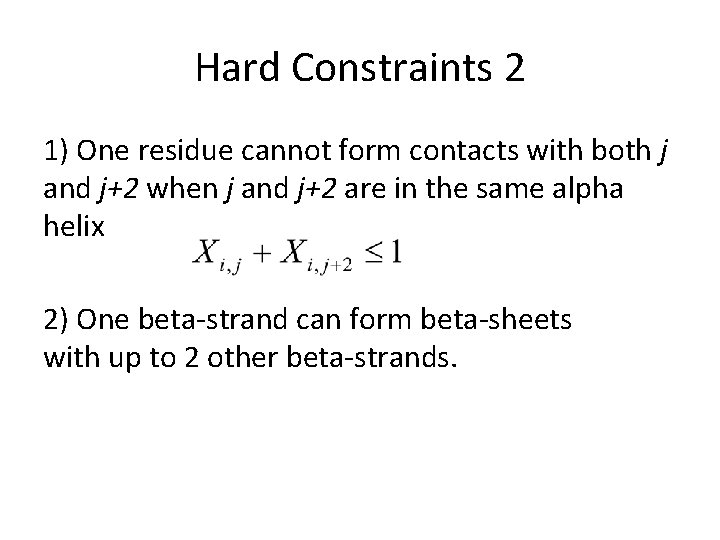
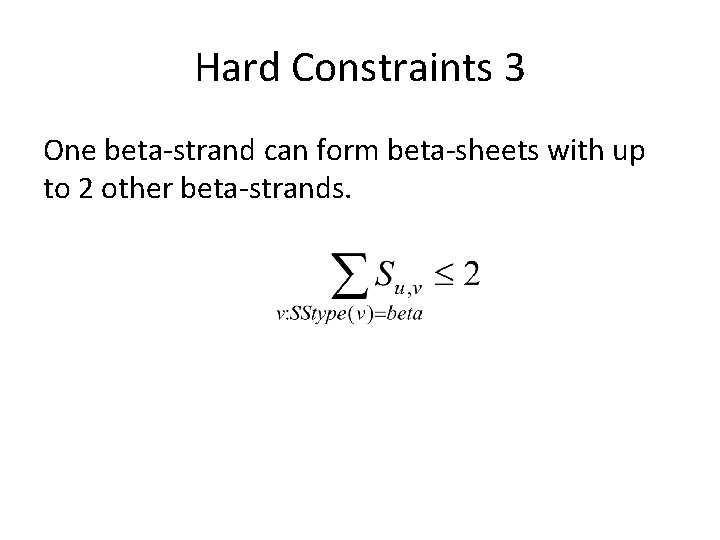
Hard Constraints 2 1) One residue cannot form contacts with both j and j+2 when j and j+2 are in the same alpha helix 2) One beta-strand can form beta-sheets with up to 2 other beta-strands.

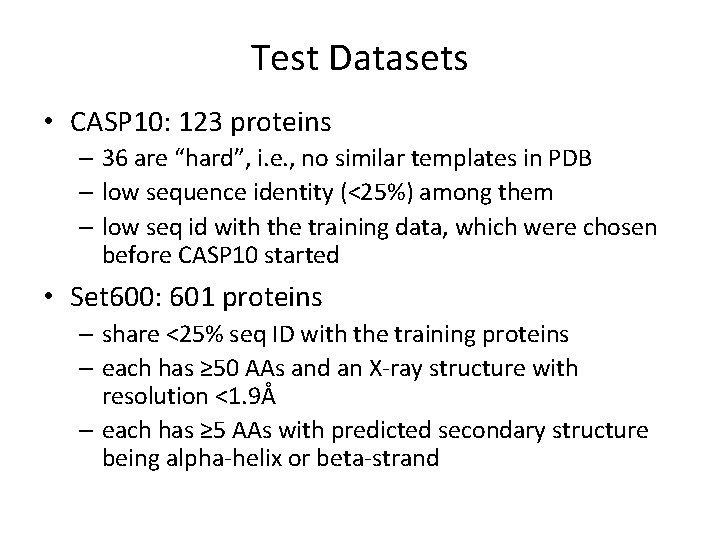
Test Datasets • CASP 10: 123 proteins – 36 are “hard”, i. e. , no similar templates in PDB – low sequence identity (<25%) among them – low seq id with the training data, which were chosen before CASP 10 started • Set 600: 601 proteins – share <25% seq ID with the training proteins – each has ≥ 50 AAs and an X-ray structure with resolution <1. 9Å – each has ≥ 5 AAs with predicted secondary structure being alpha-helix or beta-strand

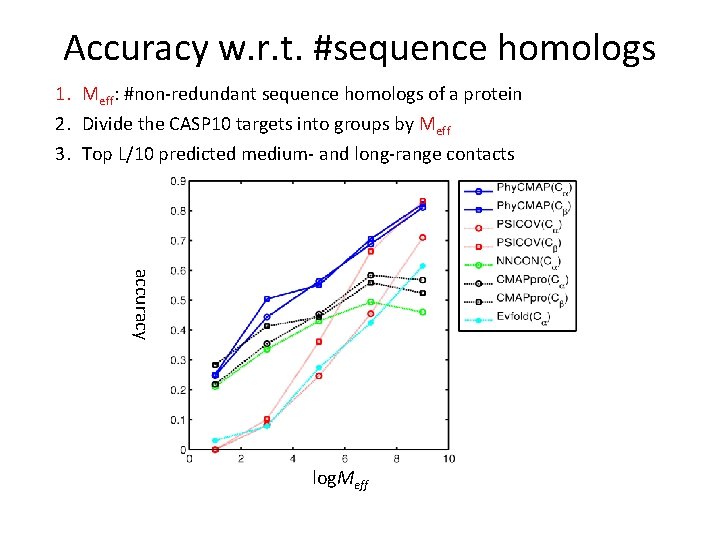
Accuracy w. r. t. #sequence homologs 1. Meff: #non-redundant sequence homologs of a protein 2. Divide the CASP 10 targets into groups by Meff 3. Top L/10 predicted medium- and long-range contacts accuracy log. Meff

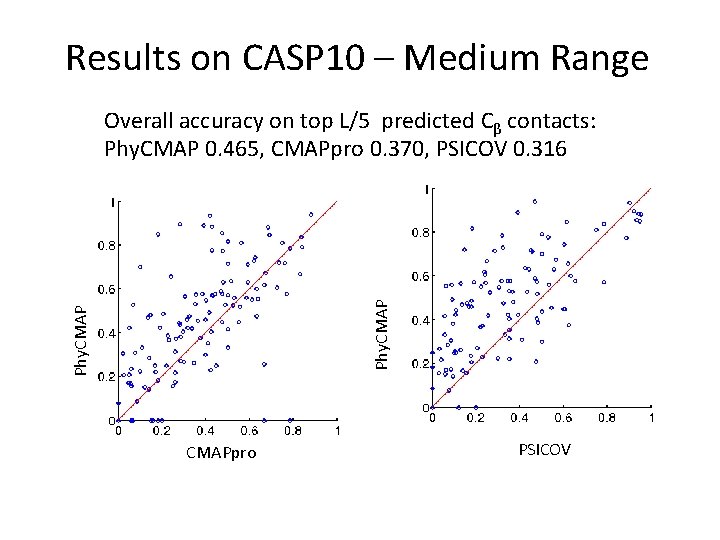
Results on CASP 10 – Medium Range Phy. CMAP Overall accuracy on top L/5 predicted Cβ contacts: Phy. CMAP 0. 465, CMAPpro 0. 370, PSICOV 0. 316 CMAPpro PSICOV

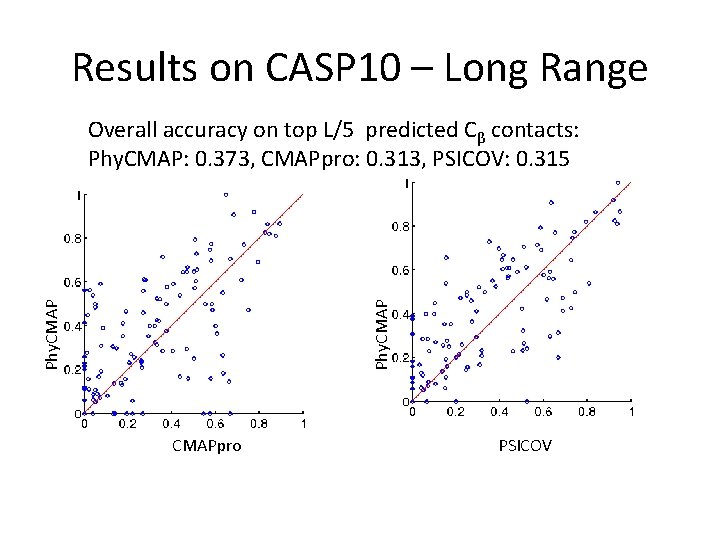
Results on CASP 10 – Long Range Phy. CMAP Overall accuracy on top L/5 predicted Cβ contacts: Phy. CMAP: 0. 373, CMAPpro: 0. 313, PSICOV: 0. 315 CMAPpro PSICOV

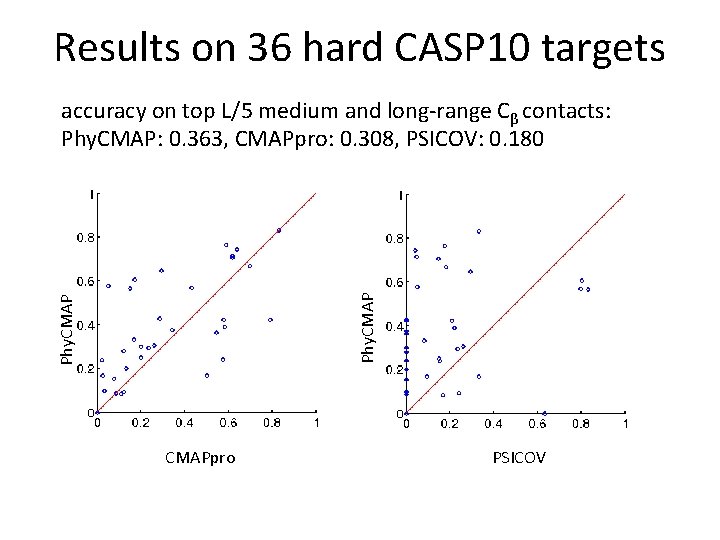
Results on 36 hard CASP 10 targets Phy. CMAP accuracy on top L/5 medium and long-range Cβ contacts: Phy. CMAP: 0. 363, CMAPpro: 0. 308, PSICOV: 0. 180 CMAPpro PSICOV

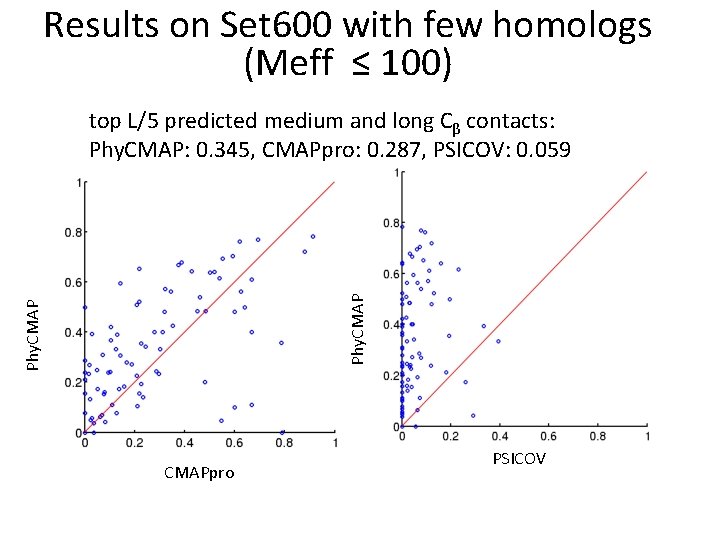
Results on Set 600 with few homologs (Meff ≤ 100) Phy. CMAP top L/5 predicted medium and long Cβ contacts: Phy. CMAP: 0. 345, CMAPpro: 0. 287, PSICOV: 0. 059 CMAPpro PSICOV

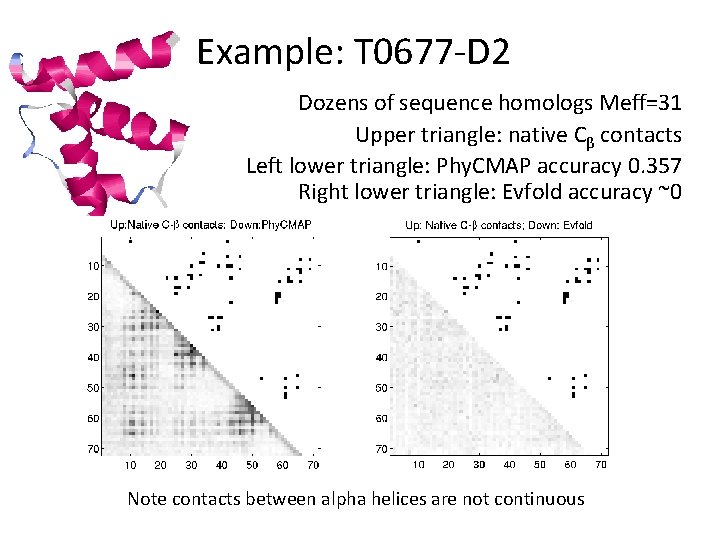
Example: T 0677 -D 2 Dozens of sequence homologs Meff=31 Upper triangle: native Cβ contacts Left lower triangle: Phy. CMAP accuracy 0. 357 Right lower triangle: Evfold accuracy ~0 Note contacts between alpha helices are not continuous

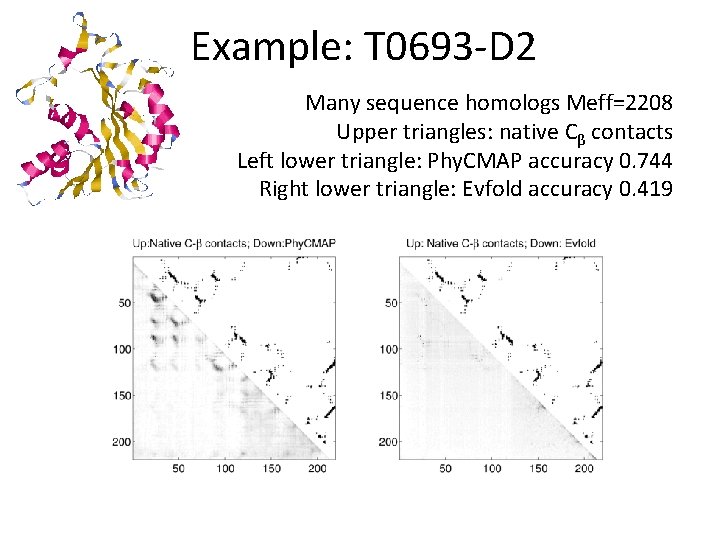
Example: T 0693 -D 2 Many sequence homologs Meff=2208 Upper triangles: native Cβ contacts Left lower triangle: Phy. CMAP accuracy 0. 744 Right lower triangle: Evfold accuracy 0. 419

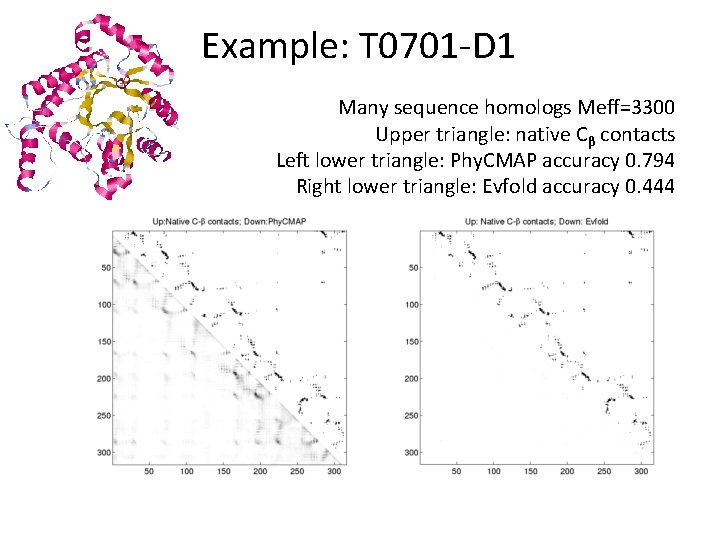
Example: T 0701 -D 1 Many sequence homologs Meff=3300 Upper triangle: native Cβ contacts Left lower triangle: Phy. CMAP accuracy 0. 794 Right lower triangle: Evfold accuracy 0. 444

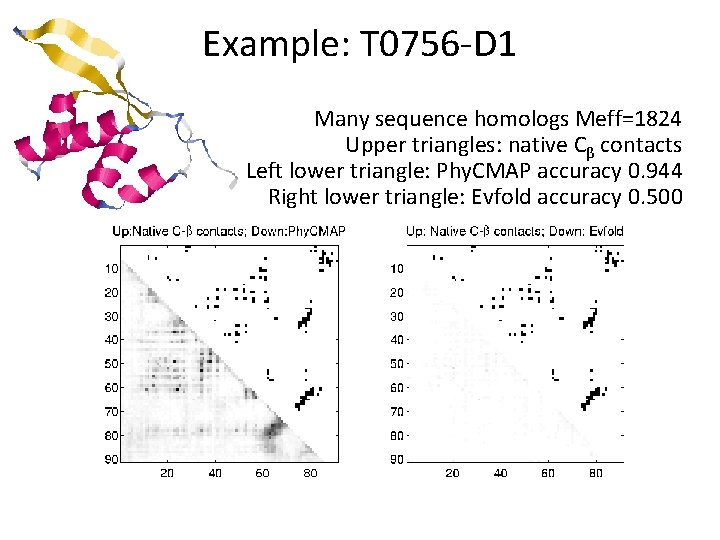
Example: T 0756 -D 1 Many sequence homologs Meff=1824 Upper triangles: native Cβ contacts Left lower triangle: Phy. CMAP accuracy 0. 944 Right lower triangle: Evfold accuracy 0. 500

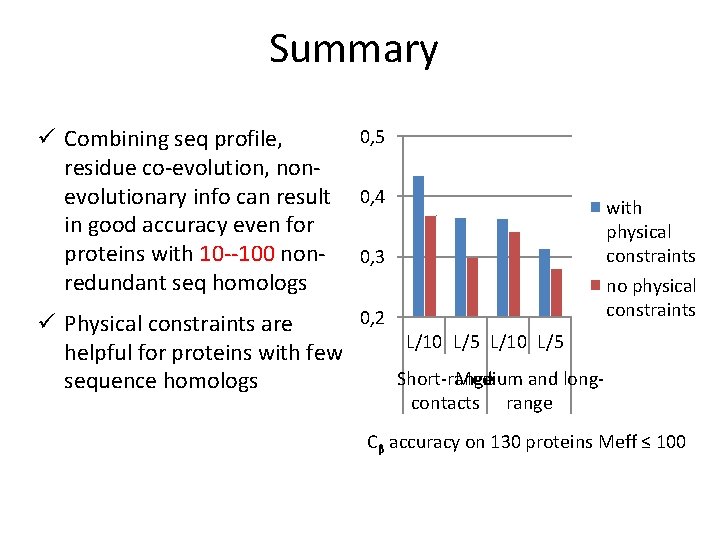
Summary ü Combining seq profile, residue co-evolution, nonevolutionary info can result in good accuracy even for proteins with 10 --100 nonredundant seq homologs 0, 5 0, 4 0, 3 0, 2 ü Physical constraints are L/10 L/5 helpful for proteins with few Short-range Medium and longsequence homologs contacts with physical constraints no physical constraints range Cβ accuracy on 130 proteins Meff ≤ 100

Acknowledgements • Student: Zhiyong Wang • Funding – NIH R 01 GM 0897532 – NSF CAREER award – Alfred P. Sloan Research Fellowship • Computational resources – University of Chicago Beagle team – Tera. Grid Web server at http: //raptorx. uchicago. edu

Protein contact Contact : Distance between two Cα or Cβ atoms < 8Å; or Distance between the closest atoms of 2 residues. 1 J 8 B short range: 6 -12 AAs apart medium range: 12 -24 AAs long range: >24 AAs apart

Why contact prediction? • Contacts describe spatial and functional relationship of residues • Contains key information for 3 D structure • Useful for protein structure prediction • Used for protein structure alignment and classification

Contrastive Mutual Information (CMI) removes local background, by measuring the MI difference between one pair of residues and neighboring pairs.

Integer Linear Programming • Objective function: • g(R): penalty for violation of physical constraints Variables Explanations Xi, j equal to 1 if there is a contact between two residues i and j. APu, v equal to 1 if two beta-strands u and v form an anti-parallel beta-sheet. Pu, v equal to 1 if two beta-strands u and v form a parallel beta-sheet. Su, v equal to 1 if two beta-strands u and v form a beta-sheet. Tu, v equal to 1 if there is an alpha-bridge between two helices u and v. Rr a non-negative integral relaxation variable of the rth soft constraint.

Hard Constraints 3 One beta-strand can form beta-sheets with up to 2 other beta-strands.

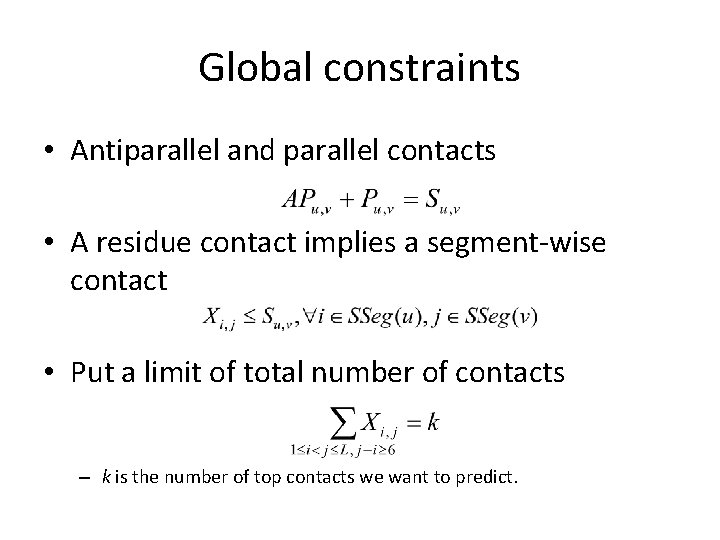
Global constraints • Antiparallel and parallel contacts • A residue contact implies a segment-wise contact • Put a limit of total number of contacts – k is the number of top contacts we want to predict.

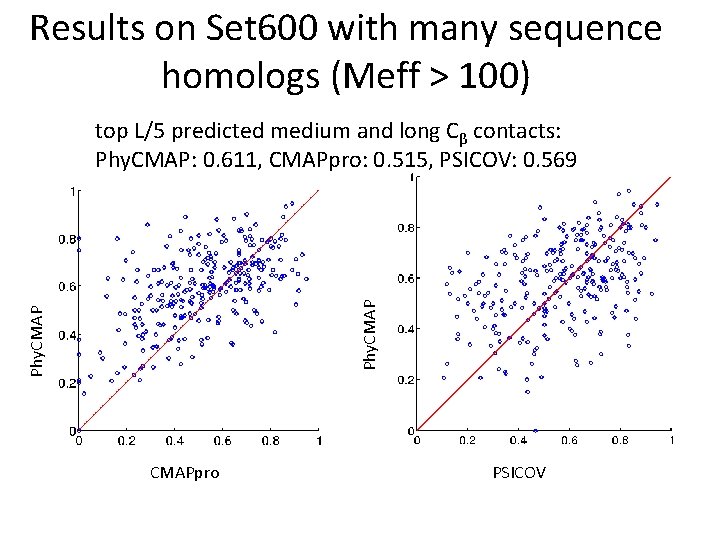
Results on Set 600 with many sequence homologs (Meff > 100) Phy. CMAP top L/5 predicted medium and long Cβ contacts: Phy. CMAP: 0. 611, CMAPpro: 0. 515, PSICOV: 0. 569 CMAPpro PSICOV

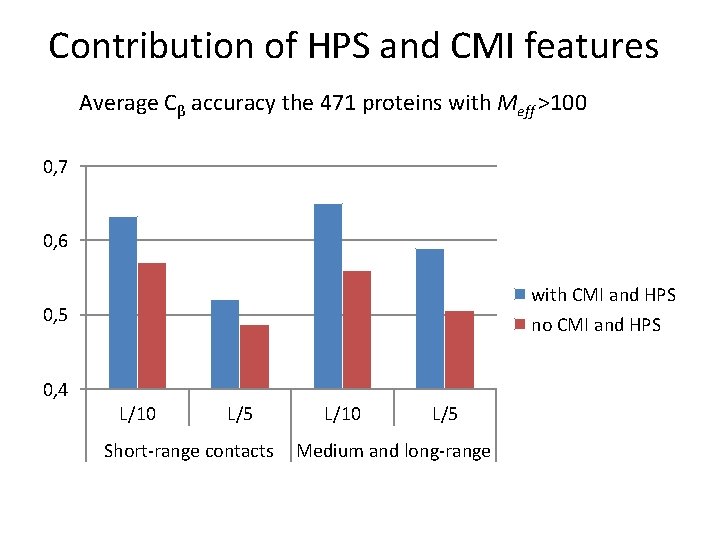
Contribution of HPS and CMI features Average Cβ accuracy the 471 proteins with Meff >100 0, 7 0, 6 with CMI and HPS no CMI and HPS 0, 5 0, 4 L/10 L/5 Short-range contacts L/10 L/5 Medium and long-range

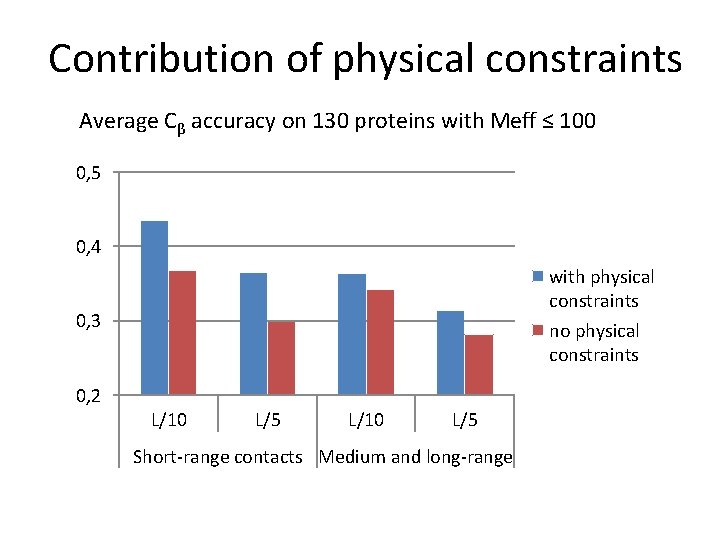
Contribution of physical constraints Average Cβ accuracy on 130 proteins with Meff ≤ 100 0, 5 0, 4 with physical constraints no physical constraints 0, 3 0, 2 L/10 L/5 Short-range contacts Medium and long-range