Pharm ID A New Algorithm for Pharmacophore Identification














- Slides: 36

Pharm. ID: A New Algorithm for Pharmacophore Identification Stan Young Jun Feng and Ashish Sanil NISS MPDM 3 June 2005 1

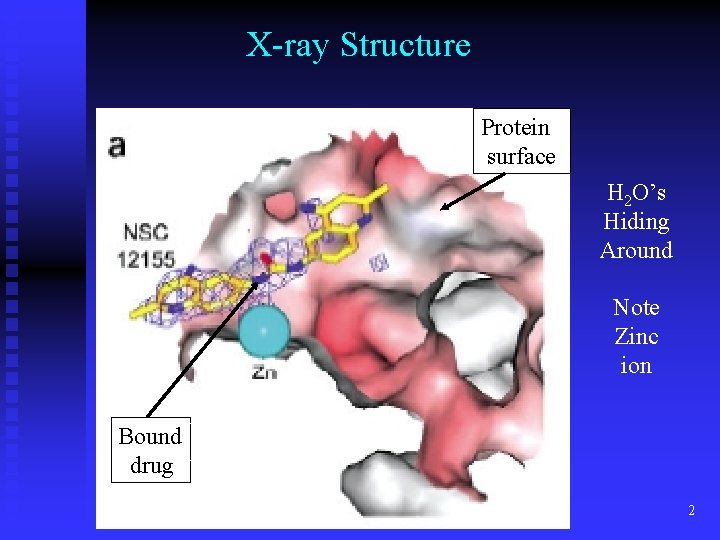
X-ray Structure Protein surface H 2 O’s Hiding Around Note Zinc ion Bound Zinc drug 2

Outline n Background n Computational Procedure and Algorithm n Examples n Conclusions 3

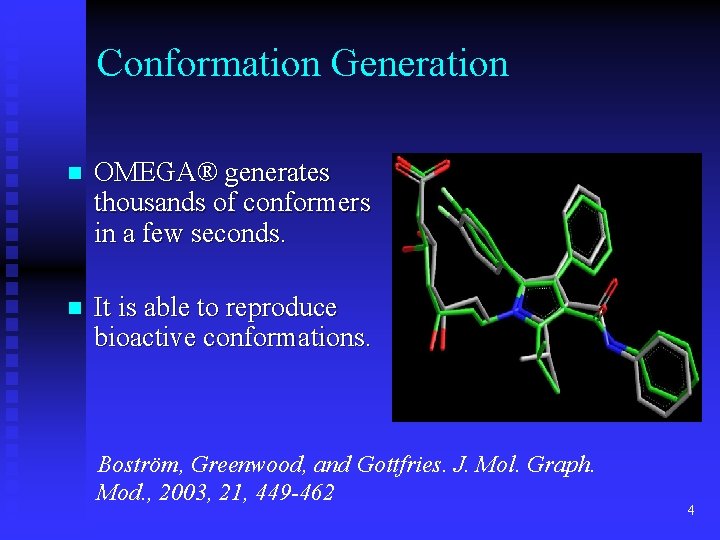
Conformation Generation n OMEGA® generates thousands of conformers in a few seconds. n It is able to reproduce bioactive conformations. Boström, Greenwood, and Gottfries. J. Mol. Graph. Mod. , 2003, 21, 449 -462 4

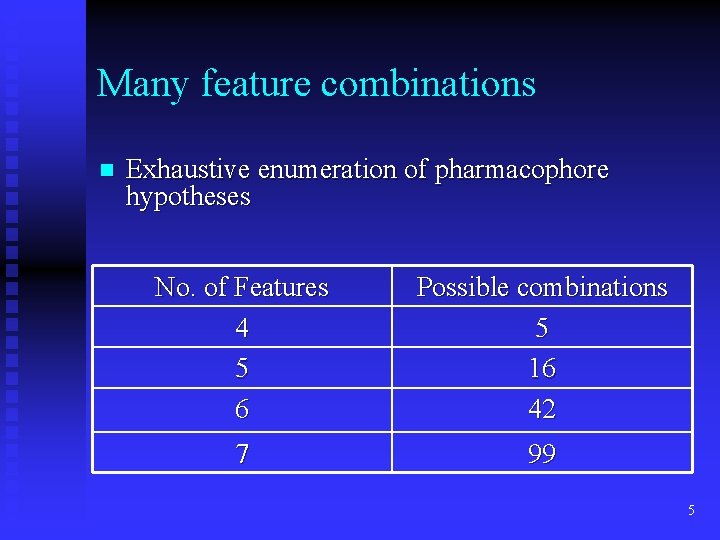
Many feature combinations n Exhaustive enumeration of pharmacophore hypotheses No. of Features 4 5 6 Possible combinations 5 16 42 7 99 5

Pharmacophore Identification n Active molecules are known, receptor unknown. Assume that all molecules bind in a common manner to the biological target. Difficulties: u Conformational flexibility u Many different combinations of pharmacophoric groups Two very large search spaces: conformations and feature combinations. 6

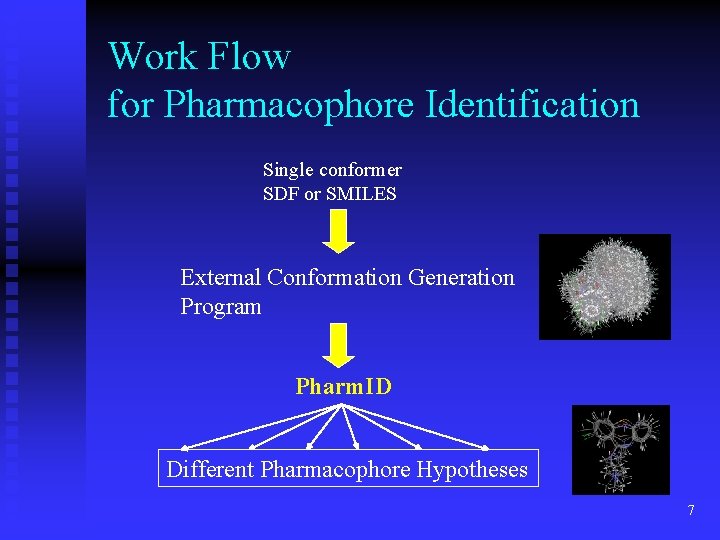
Work Flow for Pharmacophore Identification Single conformer SDF or SMILES External Conformation Generation Program Pharm. ID Different Pharmacophore Hypotheses 7

Our Strategy To superimpose the molecules in 3 D, we first align the bit string for each conformer in 1 D. n Ideally, the important features and best conformers will be picked out at the same time. n Our search is a many to one, not many to many! n 8

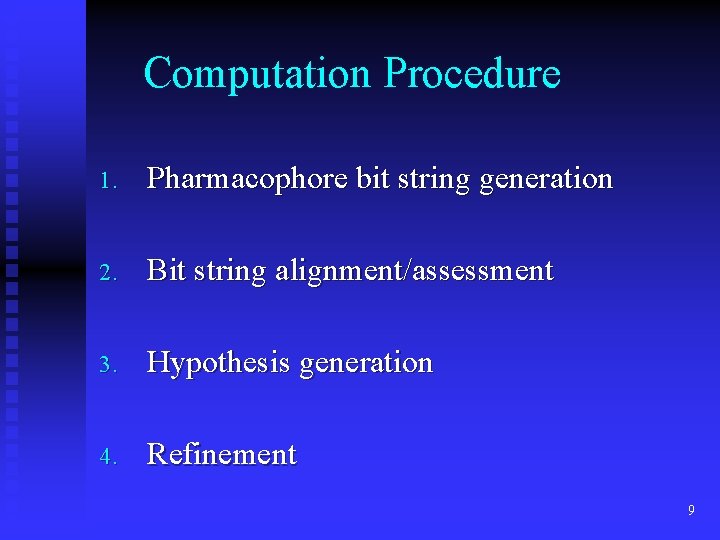
Computation Procedure 1. Pharmacophore bit string generation 2. Bit string alignment/assessment 3. Hypothesis generation 4. Refinement 9

Feature Definition n Predefined pharmacophore features: HD : Hydrogen Bond Donor HA : Hydrogen Bond Acceptor POS: Positive Charge Center NEG: Negative Charge Center ARC: Aromatic Center HYP: Hydrophobic Center n User defined groups: Any functional groups can be defined using Daylight® SMART strings. 10

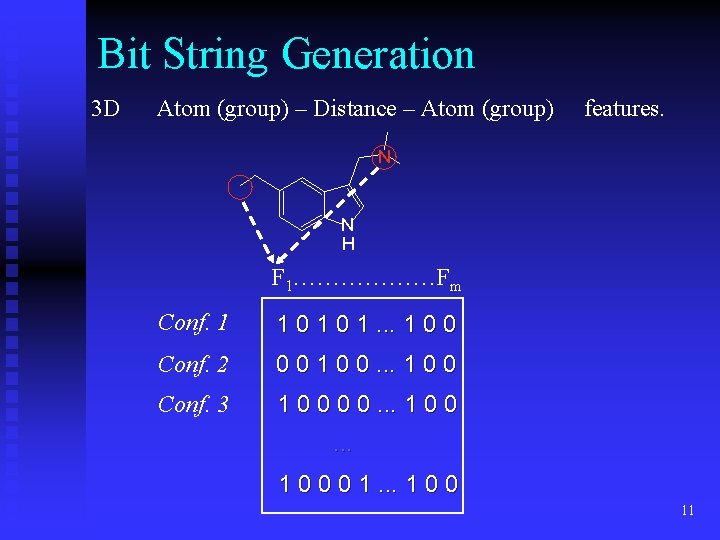
Bit String Generation 3 D Atom (group) – Distance – Atom (group) features. N N H F 1………………Fm Conf. 1 1 0 1. . . 1 0 0 Conf. 2 0 0 1 0 0. . . 1 0 0 Conf. 3 1 0 0. . . 1 0 0 0 1. . . 1 0 0 11

Definition of Distance Bins homogeneous non-overlapped. 0 -1, 1 -2, 2 -3, 4 -5, 5 -6, 6 -7, 7 -8, 8 -9, 9 -10, 11 -12, 12 Å and above. n heterogeneous non-overlapped. 1 -2, 2 -5, 5 -8, 8 -12, 13 Å and above. n Overlapped. 1 -3, 2 -4, 3 -5, 4 -6, 5 -7, 6 -8, 7 -9, 8 -10, 9 -11, 10 -12 Å. n 12

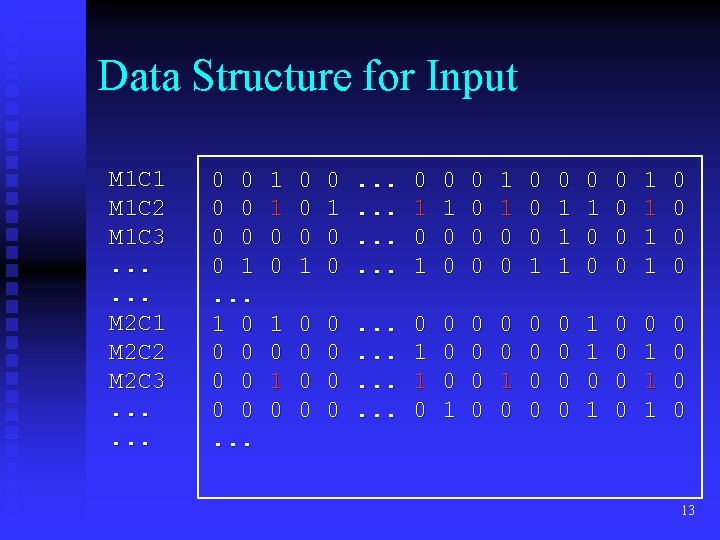
Data Structure for Input M 1 C 1 M 1 C 2 M 1 C 3. . . M 2 C 1 M 2 C 2 M 2 C 3. . . 0 0 0 0 1. . . 1 0 0 0 0. . . 1 1 0 0 0 1 0 0 . . . 0 1 0 1 0 0 0 1 1 1 0 0 0 0 1 1 0 0 0 0 0 . . . 0 1 1 0 0 0 1 0 0 0 0 0 1 1 1 0 0 13

The Trick If you know the correct conformation for each molecule, then it is relatively easy to identify the key features. If you know the correct features and distances, then it is easy to identify the correct conformation. Guess one, predict the other, iterate. 14

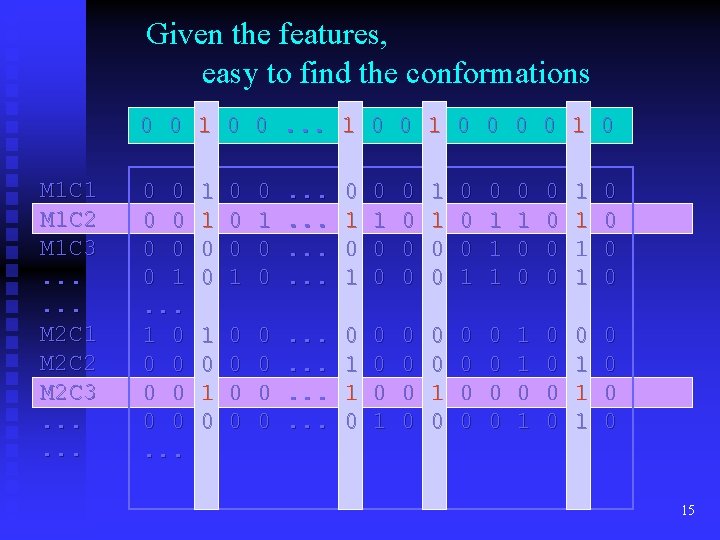
Given the features, easy to find the conformations 0 0 1 0 0. . . 1 0 0 0 0 1 0 M 1 C 1 M 1 C 2 M 1 C 3. . . M 2 C 1 M 2 C 2 M 2 C 3. . . 0 0 0 0 1. . . 1 0 0 0 0. . . 1 1 0 0 0 1 0 0 . . . 0 1 0 1 0 0 0 1 1 1 0 0 0 0 1 1 0 0 0 0 0 . . . 0 1 1 0 0 0 1 0 0 0 0 0 1 1 1 0 0 15

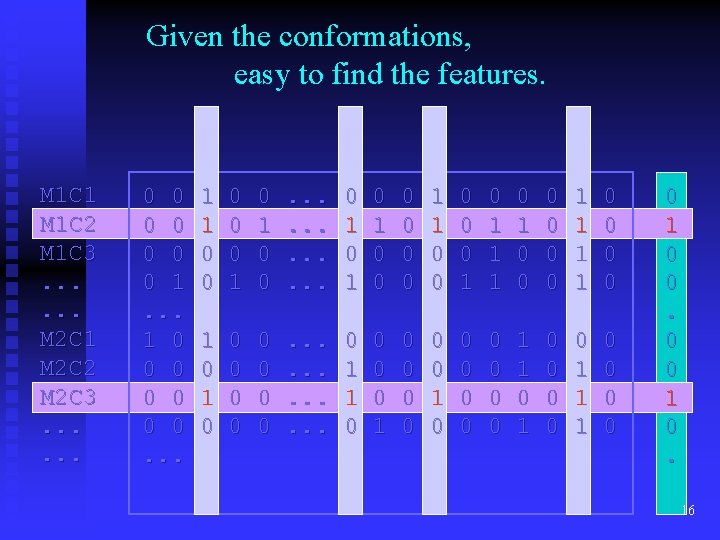
Given the conformations, easy to find the features. M 1 C 1 M 1 C 2 M 1 C 3. . . M 2 C 1 M 2 C 2 M 2 C 3. . . 0 0 0 0 1. . . 1 0 0 0 0. . . 1 1 0 0 0 1 0 0 . . . 0 1 0 1 0 0 0 1 1 1 0 0 0 0 1 1 0 0 0 0 0 . . . 0 1 1 0 0 0 1 0 0 0 0 0 1 1 1 0 0 0 1 0 0 1 0. 16

Bioinformatics Motif Finding using Gibbs Sampling. 1. 2. 3. 4. 5. Remove one sequence. Randomly select one position for each sequence. Calculate probabilities for all positions for the motif “window”. Using the “window” compute probabilities for removed sequence motif position. Repeat the above steps for all sequences until converged. This will be easier to see with pictures. 17

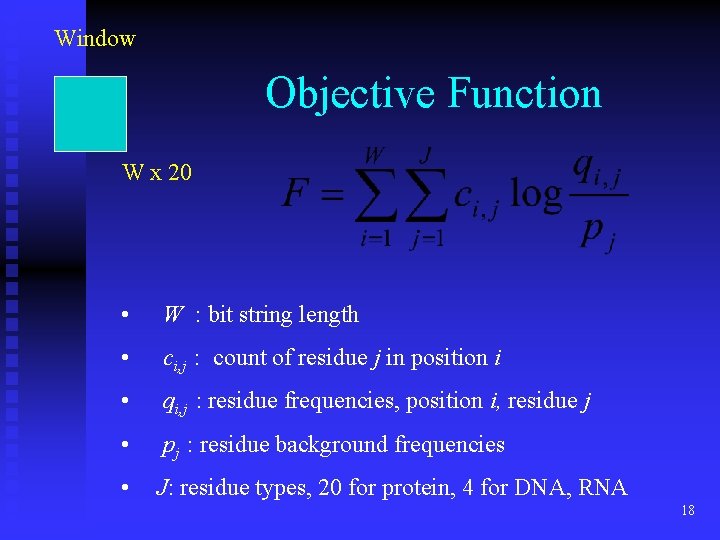
Window Objective Function W x 20 • W : bit string length • ci, j : count of residue j in position i • qi, j : residue frequencies, position i, residue j • pj : residue background frequencies • J: residue types, 20 for protein, 4 for DNA, RNA 18

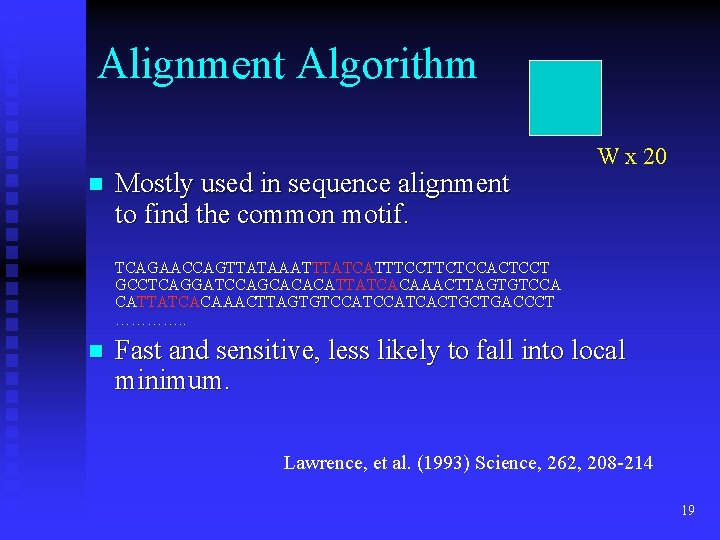
Alignment Algorithm n Mostly used in sequence alignment to find the common motif. W x 20 TCAGAACCAGTTATAAATTTATCATTTCCTTCTCCACTCCT GCCTCAGGATCCAGCACACATTATCACAAACTTAGTGTCCATCACTGCTGACCCT …………. . n Fast and sensitive, less likely to fall into local minimum. Lawrence, et al. (1993) Science, 262, 208 -214 19

Pharm. ID Algorithm using Gibbs. 1. 2. 3. 4. 5. Remove one compound. Start with a random conformer for other compounds. Calculate probabilities for feature importance. Compute conformation probabilities for omitted compound. Repeat steps 1 -4 until converges. Again, pictures will make this clear. 20

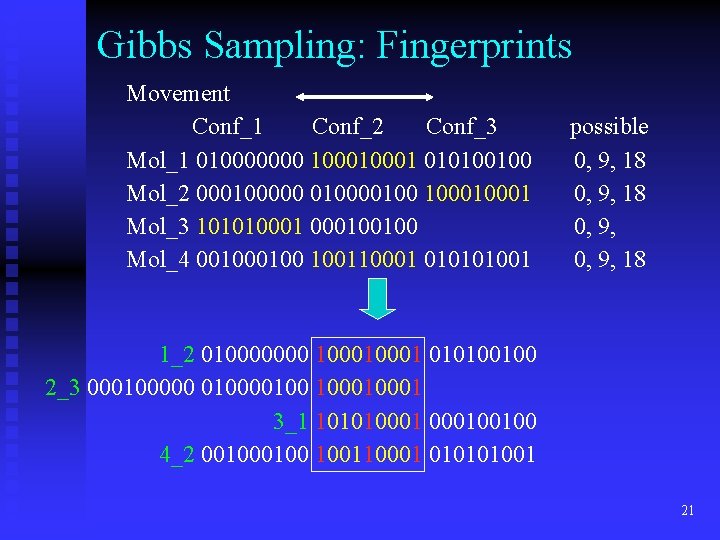
Gibbs Sampling: Fingerprints Movement Conf_1 Conf_2 Conf_3 Mol_1 010000000 10001 010100100 Mol_2 000100000 010000100 10001 Mol_3 10101000100100 Mol_4 00100 100110001 010101001 possible 0, 9, 18 1_2 010000000 10001 010100100 2_3 000100000 010000100 10001 3_1 10101000100100 4_2 00100 100110001 010101001 21

Bit String Alignment n Only 2 residue types (0, 1) n Rigid molecules that have only 1 or a few conformers can speed up the alignment and help to determine the best set of features. 22

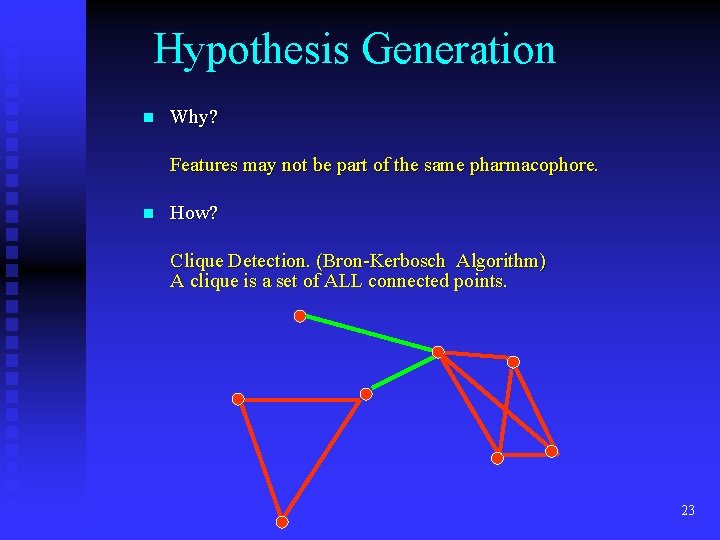
Hypothesis Generation n Why? Features may not be part of the same pharmacophore. n How? Clique Detection. (Bron-Kerbosch Algorithm) A clique is a set of ALL connected points. 23

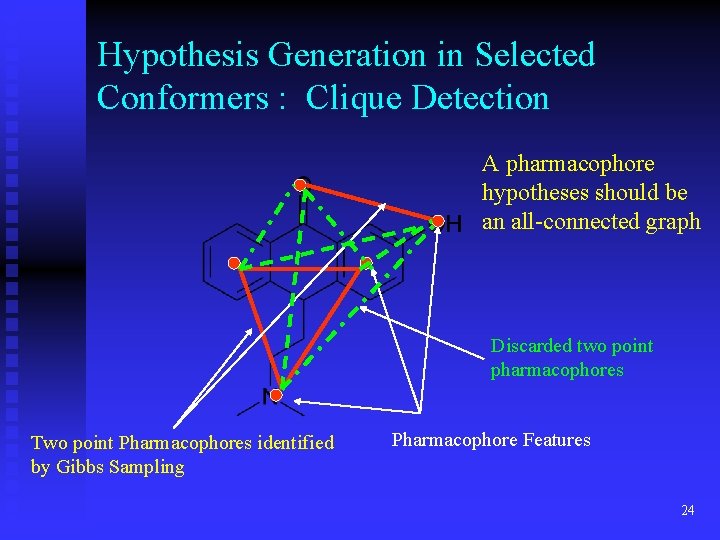
Hypothesis Generation in Selected Conformers : Clique Detection A pharmacophore hypotheses should be an all-connected graph Discarded two point pharmacophores Two point Pharmacophores identified by Gibbs Sampling Pharmacophore Features 24

Hypothesis Generation: Output n n Pharmacophore 1 Members: 1 2 3 5 …(Mol. ID) Features: Hydrogen Bond Donor, Hydrogen Bond Acceptor, … Pharmcophore 2 Members: 4 6 8 … Features: … … … 25

Refinement For all molecules u For all conformers t For all hypotheses generated Test each qualified conformer against each hypothesis End For If new hypothesis found Insert the new hypothesis into the list End For n 26

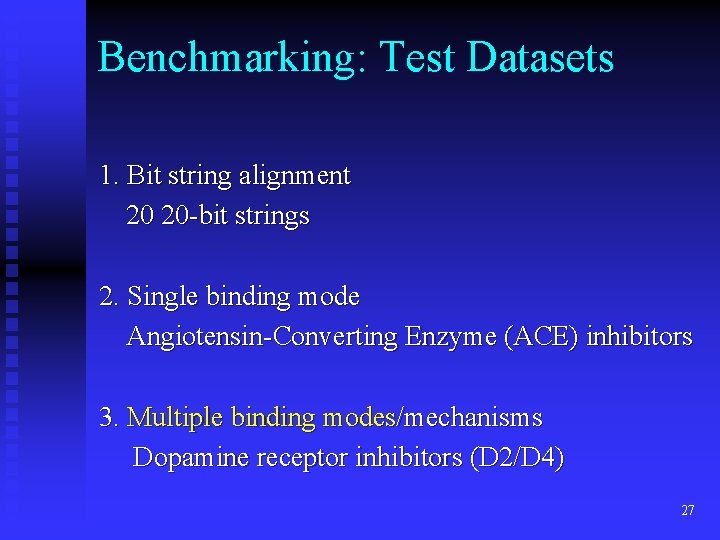
Benchmarking: Test Datasets 1. Bit string alignment 20 20 -bit strings 2. Single binding mode Angiotensin-Converting Enzyme (ACE) inhibitors 3. Multiple binding modes/mechanisms Dopamine receptor inhibitors (D 2/D 4) 27

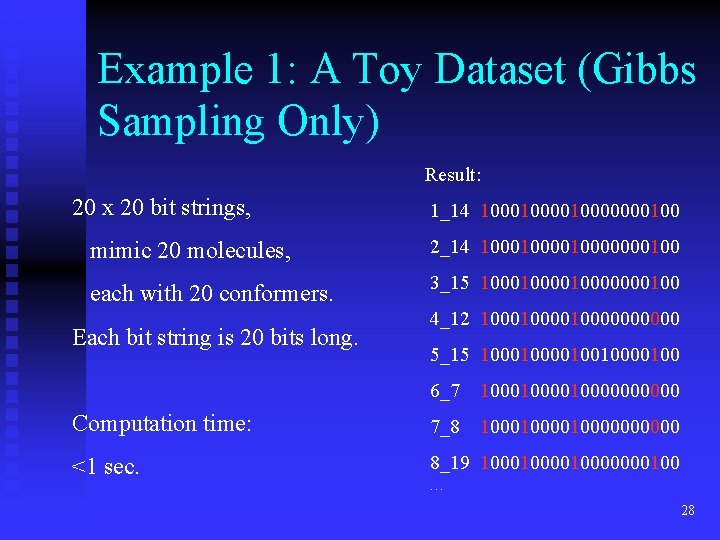
Example 1: A Toy Dataset (Gibbs Sampling Only) Result: 20 x 20 bit strings, 1_14 1000010000000100 mimic 20 molecules, 2_14 1000010000000100 each with 20 conformers. 3_15 1000010000000100 Each bit string is 20 bits long. 4_12 10000100000 5_15 10000100100 6_7 10000100000 Computation time: 7_8 10000100000 <1 sec. 8_19 1000010000000100 … 28

Example 2: ACE Inhibitors n 78 active compounds. n OMEGA® From Open. Eye® is used to generate multiple conformers. n Two RMSD cutoffs used: 2. 0 Å : 4, 613 conformers generated. 1. 0 Å : 46, 268 conformers generated. 29

ACE inhibitors Results n Using 4, 613 conformers, 55/78 molecules contain expected pharmacophore. n Using 46, 268 conformers, 65/78 molecules contain expected pharmacophore. 30

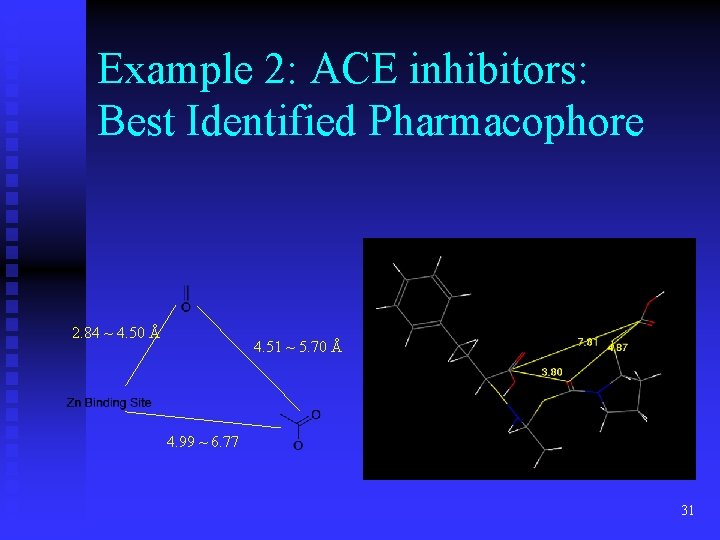
Example 2: ACE inhibitors: Best Identified Pharmacophore 2. 84 ~ 4. 50 Å 4. 51 ~ 5. 70 Å 4. 99 ~ 6. 77 31

Example 2: ACE inhibitors Other possible pharmacophore 32

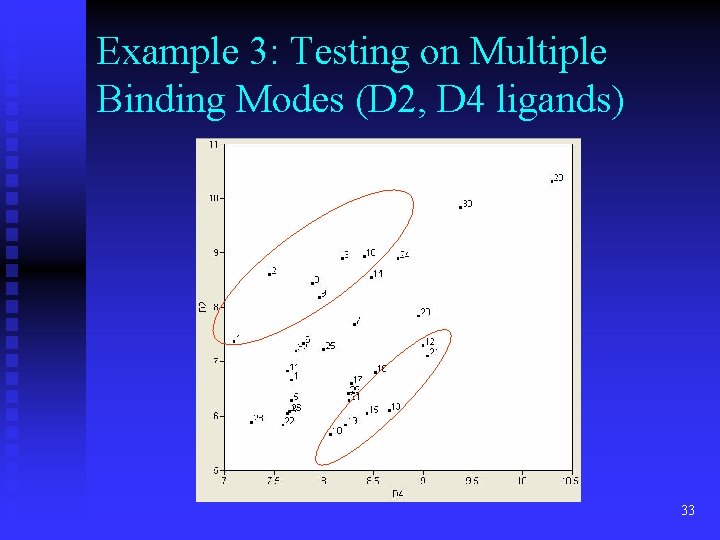
Example 3: Testing on Multiple Binding Modes (D 2, D 4 ligands) 33

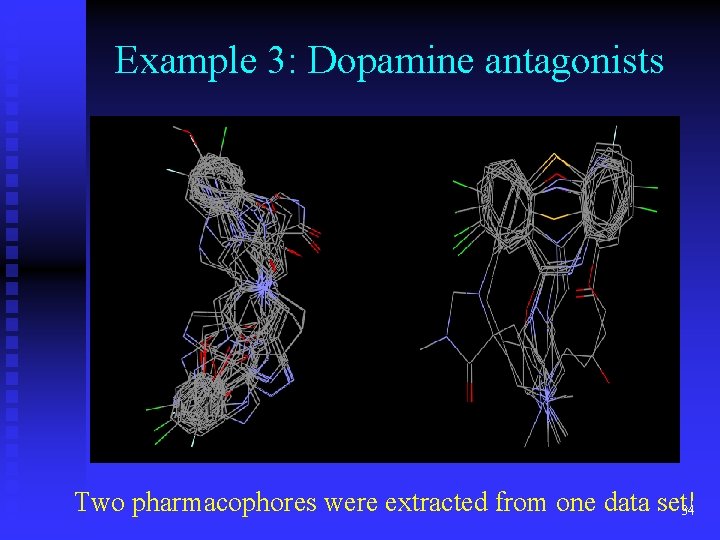
Example 3: Dopamine antagonists Two pharmacophores were extracted from one data set! 34

Conclusion n Traditional Methods: Exhaustive enumeration of pharmacophores, limited coverage of conformational space. “Many to many” limits search. n Pharm. ID: Selective enumeration of pharmacophores, better coverage of conformational space. Each search is “many to one”. 35

Acknowledgements n Coworkers Stan Young, Jun Feng, Ashish Sanil n OMEGA is a product from Open. Eye Scientific Software Inc. n Support from Hereditary Disease Foundation. Become a NISS affiliate! 36