Personalized Predictive Medicine and Genomic Clinical Trials Richard












































- Slides: 60

Personalized Predictive Medicine and Genomic Clinical Trials Richard Simon, D. Sc. Chief, Biometric Research Branch National Cancer Institute http: //brb. nci. nih. gov

Biometric Research Branch Website brb. nci. nih. gov Powerpoint presentations n Reprints n BRB-Array. Tools software n Web based Sample Size Planning n

Personalized Oncology is Here Today and Rapidly Advancing Key information is in tumor genome, not in inherited genetics n Personalization is based on limited stratification of traditional diagnostic categories, not on individual genomes n

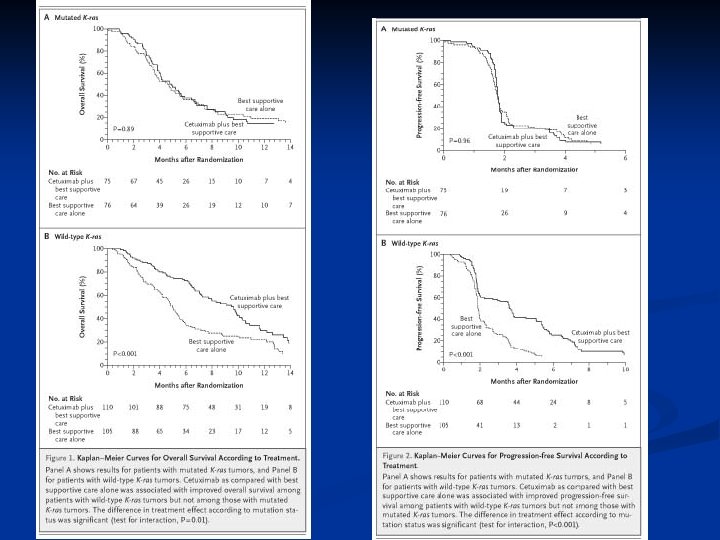
Personalized Oncology is Here Today n Estrogen receptor over-expression in breast cancer n n HER 2 amplification in breast cancer n n Low score for ER+ node - = hormonal rx KRAS in colorectal cancer n n Trastuzumab, Lapatinib Oncotype. Dx in breast cancer n n Anti-estrogens, aromatase inhibitors WT KRAS = cetuximab or panitumumab EGFR mutation or amplification in NSCLC n EGFR inhibitor

These Diagnostics Have Medical Utility They inform therapeutic decision-making leading to improved patient outcome n Tests with medical utility help patients and may reduce medical costs n Tests correlated with outcome that are not actionable may increase medical costs without helping patients n

n Developing a test and demonstrating medical utility for it is a complex multi-step process that generally requires prospective randomized clinical trials

n Although the randomized clinical trial remains of fundamental importance for predictive genomic medicine, some of the conventional wisdom of how to design and analyze rct’s requires re-examination n E. g. The concept of doing a rct of thousands of patients to answer a single question about average treatment effect for a heterogeneous target population no longer has an adequate scientific basis in oncology

Standard Approach is Based on Assumptions n Qualitative treatment by subset interactions are unlikely n i. e. if new treatment T is better than control C on average, it is better for all subsets of patients n “Costs” of over-treatment are less than “costs” of under-treatment

n Cancers of a primary site often represent a heterogeneous group of diverse molecular diseases which vary fundamentally with regard to the oncogenic mutations that cause them, n their responsiveness to specific drugs n

How Can We Develop New Drugs in a Manner More Consistent With Modern Tumor Biology and Obtain Reliable Information About What Regimens Work for What Kinds of Patients?

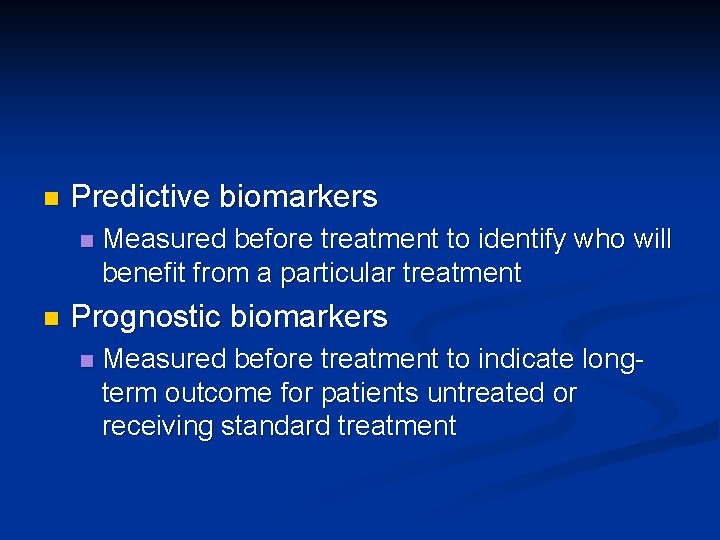
n Predictive biomarkers n n Measured before treatment to identify who will benefit from a particular treatment Prognostic biomarkers n Measured before treatment to indicate longterm outcome for patients untreated or receiving standard treatment

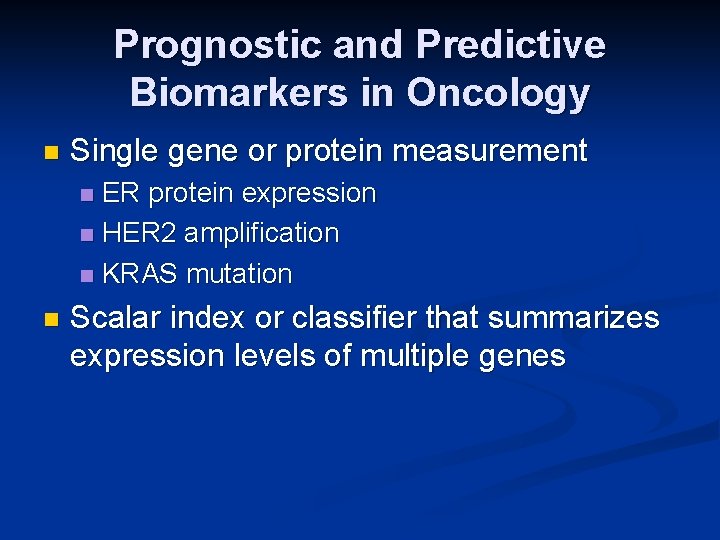
Prognostic and Predictive Biomarkers in Oncology n Single gene or protein measurement ER protein expression n HER 2 amplification n KRAS mutation n n Scalar index or classifier that summarizes expression levels of multiple genes




Prospective Co-Development of Drugs and Companion Diagnostics 1. 2. 3. Develop a completely specified genomic classifier of the patients likely to benefit from a new drug Establish analytical validity of the classifier Use the completely specified classifier to design and analyze a focused clinical trial to evaluate effectiveness of the new treatment and how it relates to the candidate biomarker

Targeted (Enrichment) Design n Restrict entry to the phase III trial based on the binary predictive classifier

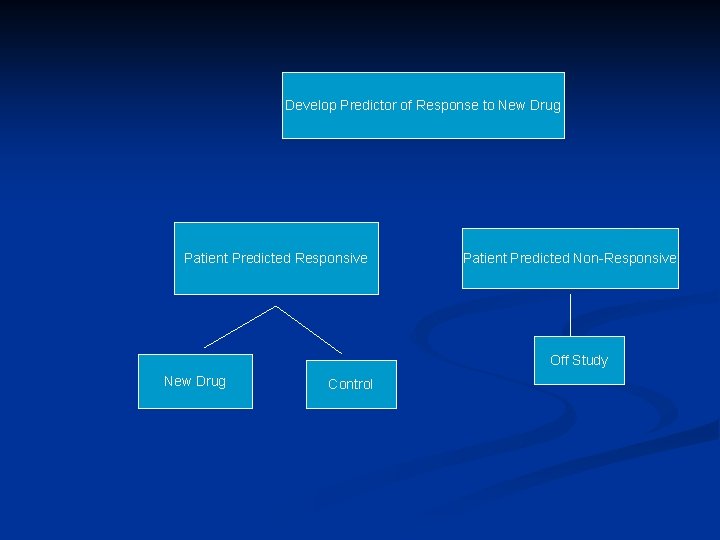
Using phase II data, develop predictor of response to new drugto New Drug Develop Predictor of Response Patient Predicted Responsive Patient Predicted Non-Responsive Off Study New Drug Control

Applicability of Targeted Design n Primarily for settings where the classifier is based on a single gene whose protein product is the target of the drug n eg trastuzumab

Evaluating the Efficiency of Targeted Design n Simon R and Maitnourim A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clinical Cancer Research 10: 6759 -63, 2004; Correction and supplement 12: 3229, 2006 Maitnourim A and Simon R. On the efficiency of targeted clinical trials. Statistics in Medicine 24: 329 -339, 2005. reprints and interactive sample size calculations at http: //linus. nci. nih. gov

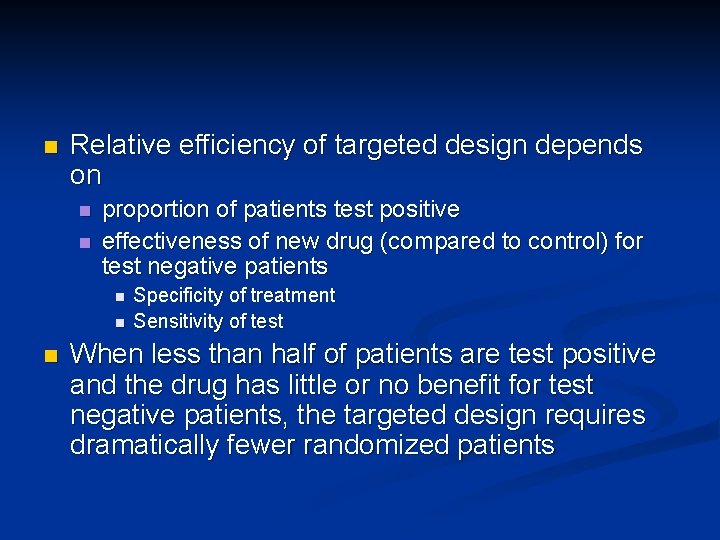
n Relative efficiency of targeted design depends on n n proportion of patients test positive effectiveness of new drug (compared to control) for test negative patients n n n Specificity of treatment Sensitivity of test When less than half of patients are test positive and the drug has little or no benefit for test negative patients, the targeted design requires dramatically fewer randomized patients

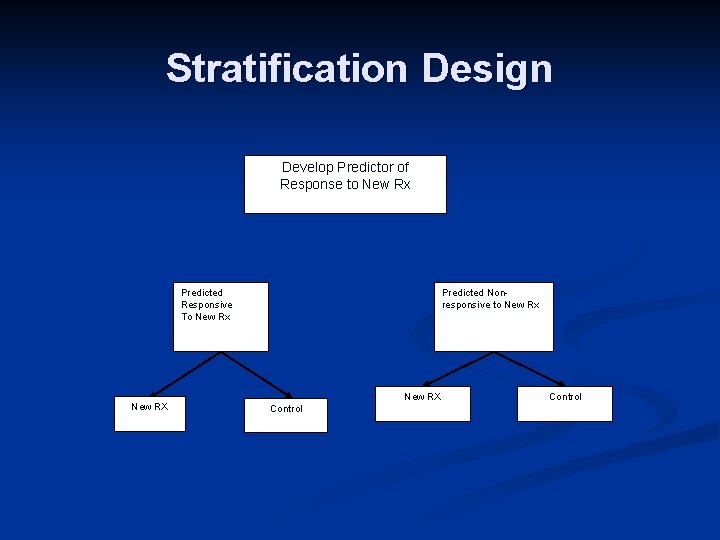
Stratification Design Develop Predictor of Response to New Rx Predicted Responsive To New Rx New RX Predicted Nonresponsive to New Rx New RX Control

n n n Do not use the test to restrict eligibility, but to structure a prospective analysis plan Having a prospective analysis plan is essential “Stratifying” (balancing) the randomization is useful to ensure that all randomized patients have tissue available but is not a substitute for a prospective analysis plan Size the study for adequate evaluation of T vs C separately by marker status The purpose of the study is to evaluate the new treatment overall and for the pre-defined subsets; not to modify or refine the classifier The purpose is not to demonstrate that repeating the classifier development process on independent data results in the same classifier

n n R Simon. Using genomics in clinical trial design, Clinical Cancer Research 14: 5984 -93, 2008 R Simon. Designs and adaptive analysis plans for pivotal clinical trials of therapeutics and companion diagnostics, Expert Opinion in Medical Diagnostics 2: 721 -29, 2008

Analysis Plan B (Limited confidence in test) n Compare the new drug to the control overall for all patients ignoring the classifier. n n If poverall ≤ 0. 03 claim effectiveness for the eligible population as a whole Otherwise perform a single subset analysis evaluating the new drug in the classifier + patients n If psubset ≤ 0. 02 claim effectiveness for the classifier + patients.

Sample size for Analysis Plan B n n To have 90% power for detecting uniform 33% reduction in overall hazard at 3% two-sided level requires 297 events (instead of 263 for similar power at 5% level) If 25% of patients are positive, then when there are 297 total events there will be approximately 75 events in positive patients n n 75 events provides 75% power for detecting 50% reduction in hazard at 2% two-sided significance level By delaying evaluation in test positive patients, 80% power is achieved with 84 events and 90% power with 109 events

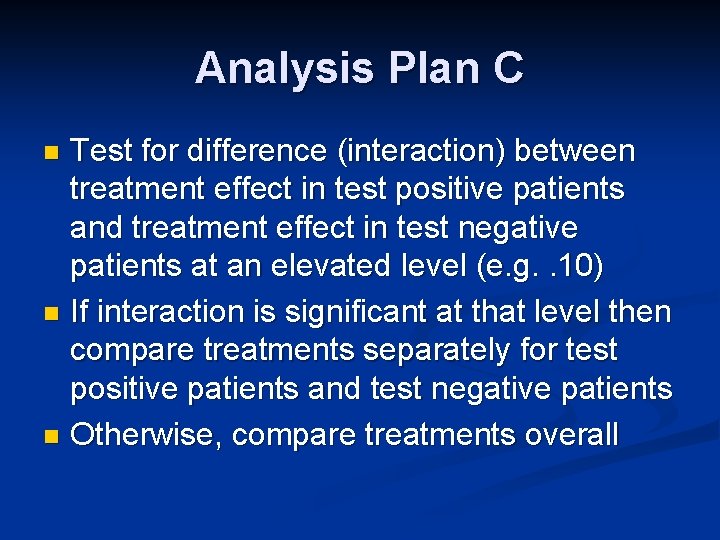
Analysis Plan C Test for difference (interaction) between treatment effect in test positive patients and treatment effect in test negative patients at an elevated level (e. g. . 10) n If interaction is significant at that level then compare treatments separately for test positive patients and test negative patients n Otherwise, compare treatments overall n

Sample Size Planning for Analysis Plan C 88 events in test + patients needed to detect 50% reduction in hazard at 5% twosided significance level with 90% power n If 25% of patients are positive, when there are 88 events in positive patients there will be about 264 events in negative patients n n 264 events provides 90% power for detecting 33% reduction in hazard at 5% two-sided significance level

Does the RCT Need to Be Significant Overall for the T vs C Treatment Comparison? n No n That requirement has been traditionally used to protect against data dredging. It is inappropriate for focused trials of a treatment with a companion test.

Web Based Software for Planning Clinical Trials of Treatments with a Candidate Predictive Biomarker n http: //brb. nci. nih. gov



n It is difficult to have the right single completely defined predictive biomarker identified analytically validated by the time the pivotal trial of a new drug is ready to start accrual n n n Changes in the way we do phase II trials Adaptive methods for the refinement and evaluation of predictive biomarkers in the pivotal trials in a nonexploratory manner Use of archived tissues in focused “prospectiveretrospective” designs based on randomized pivotal trials

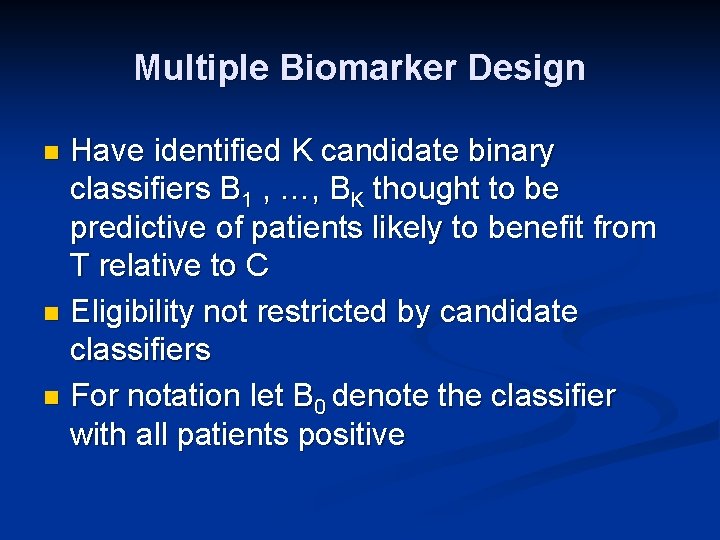
Multiple Biomarker Design Have identified K candidate binary classifiers B 1 , …, BK thought to be predictive of patients likely to benefit from T relative to C n Eligibility not restricted by candidate classifiers n For notation let B 0 denote the classifier with all patients positive n

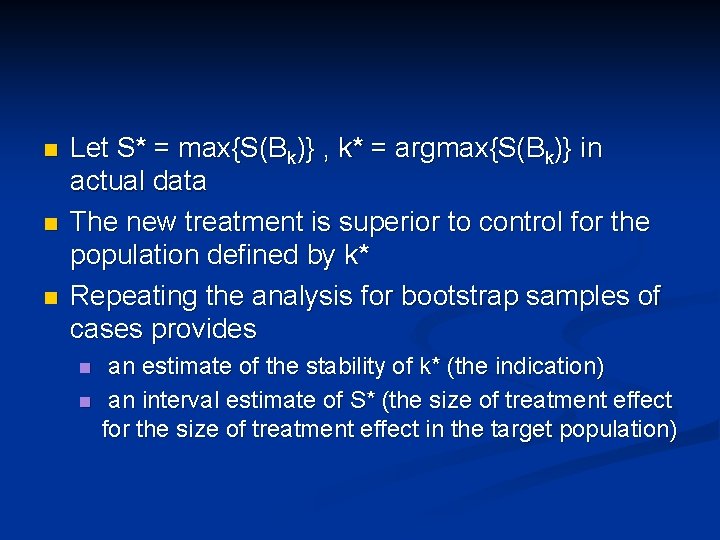
n Test T vs C restricted to patients positive for Bk for k=0, 1, …, K n n n Let S(Bk) be log partial likelihood ratio statistic for treatment effect in patients positive for Bk (k=1, …, K) Let S* = max{S(Bk)} , k* = argmax{S(Bk)} For a global test of significance n n Compute null distribution of S* by permuting treatment labels If the data value of S* is significant at 0. 05 level, then claim effectiveness of T for patients positive for Bk*

n n n Let S* = max{S(Bk)} , k* = argmax{S(Bk)} in actual data The new treatment is superior to control for the population defined by k* Repeating the analysis for bootstrap samples of cases provides n n an estimate of the stability of k* (the indication) an interval estimate of S* (the size of treatment effect for the size of treatment effect in the target population)


Adaptive Signature Design Boris Freidlin and Richard Simon Clinical Cancer Research 11: 7872 -8, 2005

Adaptive Signature Design End of Trial Analysis n Compare E to C for all patients at significance level α 0 (eg 0. 04) If overall H 0 is rejected, then claim effectiveness of E for eligible patients n Otherwise n

n Otherwise: n n Using only the first half of patients accrued during the trial, develop a binary classifier that predicts the subset of patients most likely to benefit from the new treatment T compared to control C Compare T to C for patients accrued in second stage who are predicted responsive to T based on classifier n n Perform test at significance level 1 - α 0 (eg 0. 01) If H 0 is rejected, claim effectiveness of T for subset defined by classifier

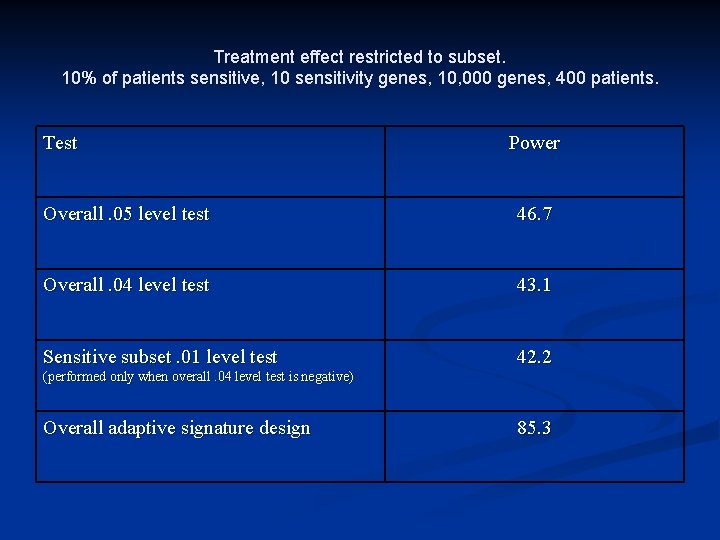
Treatment effect restricted to subset. 10% of patients sensitive, 10 sensitivity genes, 10, 000 genes, 400 patients. Test Power Overall. 05 level test 46. 7 Overall. 04 level test 43. 1 Sensitive subset. 01 level test 42. 2 (performed only when overall. 04 level test is negative) Overall adaptive signature design 85. 3

Cross-Validated Adaptive Signature Design Freidlin B, Jiang W, Simon R Clinical Cancer Research 16(2) 2010

Prediction Based Analysis of Clinical Trials Using cross-validation we can evaluate our methods for analysis of clinical trials, including complex subset analysis algorithms, in terms of their effect on improving patient outcome via informing therapeutic decision making n This approach can be used with any set of candidate predictor variables n

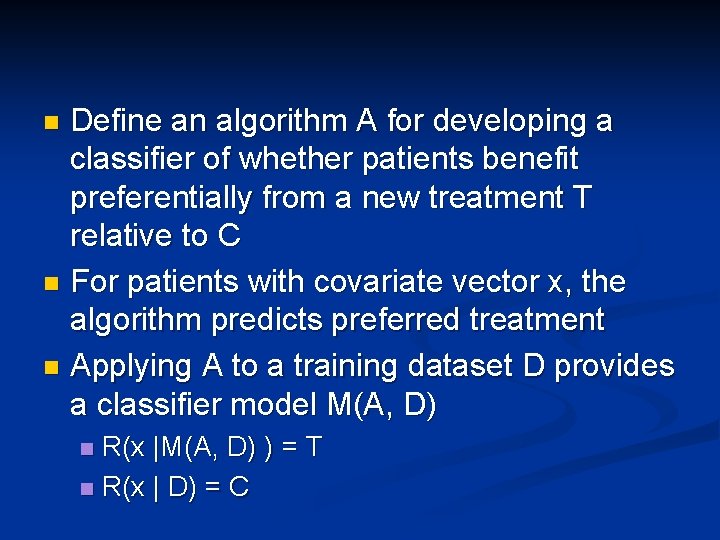
Define an algorithm A for developing a classifier of whether patients benefit preferentially from a new treatment T relative to C n For patients with covariate vector x, the algorithm predicts preferred treatment n Applying A to a training dataset D provides a classifier model M(A, D) n R(x |M(A, D) ) = T n R(x | D) = C n

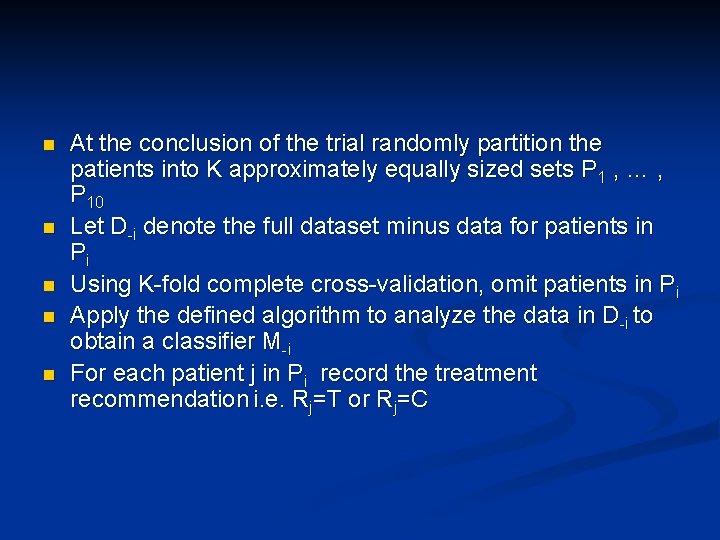
n n n At the conclusion of the trial randomly partition the patients into K approximately equally sized sets P 1 , … , P 10 Let D-i denote the full dataset minus data for patients in Pi Using K-fold complete cross-validation, omit patients in Pi Apply the defined algorithm to analyze the data in D-i to obtain a classifier M-i For each patient j in Pi record the treatment recommendation i. e. Rj=T or Rj=C

Repeat the above for all K loops of the cross-validation n All patients have been classified as what their optimal treatment is predicted to be n

n n Let ST denote the set of patients for whom treatment T is predicted optimal i. e. ST = {i : Rj=T} Compare outcomes for patients in S who actually received T to those in S who actually received C n n n Let z. T= standardized log-rank statistic Let SC denote the set of patients for whom treatment C is predicted optimal i. e. SC = {i : Rj=C} Compare outcomes for patients in SC who actually received T to those in S who actually received C n Let z. C = standardized log-rank statistic

Test of Significance for Effectiveness of T vs C n Compute statistical significance of z. T and z. C by randomly permuting treatment labels and repeating the entire procedure n Do this 1000 or more times to generate the permutation null distribution of treatment effect for the patients in each subset

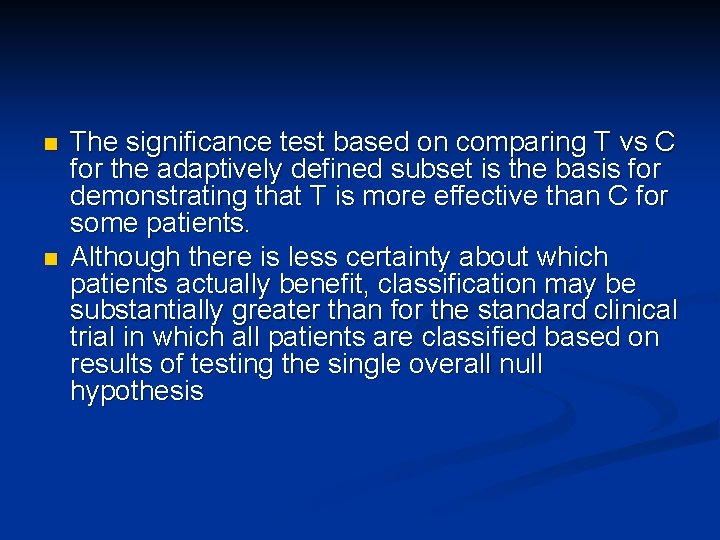
n n The significance test based on comparing T vs C for the adaptively defined subset is the basis for demonstrating that T is more effective than C for some patients. Although there is less certainty about which patients actually benefit, classification may be substantially greater than for the standard clinical trial in which all patients are classified based on results of testing the single overall null hypothesis

70% Response to T in Sensitive Patients 25% Response to T Otherwise 25% Response to C 20% Patients Sensitive ASD CV-ASD Overall 0. 05 Test 0. 486 0. 503 Overall 0. 04 Test 0. 452 0. 471 Sensitive Subset 0. 01 Test 0. 207 0. 588 Overall Power 0. 525 0. 731


Expected 5 -Year DFS Using New Algorithm n Let S(T) = observed 5 -year DFS for patients in ST who received treatment T n n Let S(C) = observed K-year DFS for patients in SC who received treatment C n n m. T such patients m. C such patients Expected K-Year DFS using new algorithm {m. T S(T) + m. C S(C)}/{m. T + m. C} Confidence limits for this estimate can be obtained by bootstrapping the complete crossvalidation procedure

Expected 5 -Year DFS Using Standard Analysis n If the overall null hypothesis is not rejected n n Expected 5 -Year DFS is the observed 5 -year DFS in the control group If the overall null hypothesis is rejected n Expected 5 -Year DFS is the observed 5 -year DFS in T group

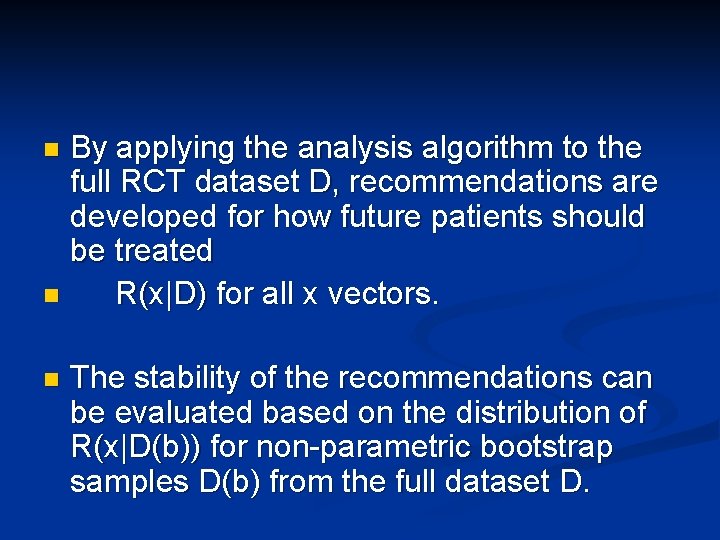
By applying the analysis algorithm to the full RCT dataset D, recommendations are developed for how future patients should be treated n R(x|D) for all x vectors. n n The stability of the recommendations can be evaluated based on the distribution of R(x|D(b)) for non-parametric bootstrap samples D(b) from the full dataset D.

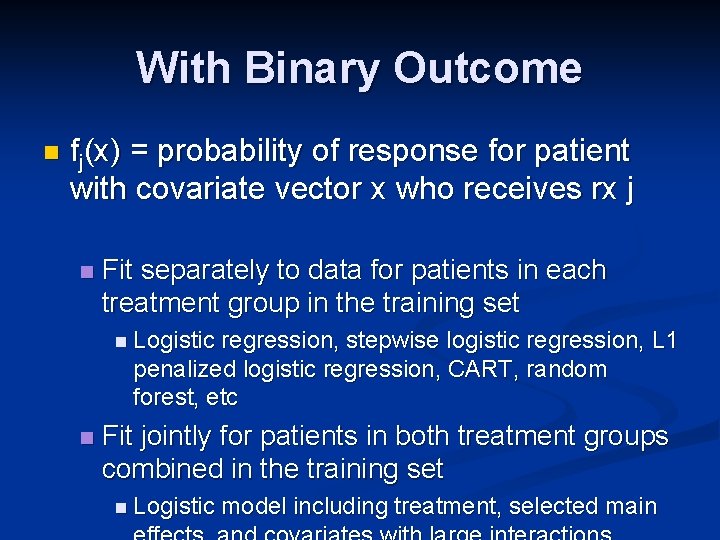
With Binary Outcome n fj(x) = probability of response for patient with covariate vector x who receives rx j n Fit separately to data for patients in each treatment group in the training set n Logistic regression, stepwise logistic regression, L 1 penalized logistic regression, CART, random forest, etc n Fit jointly for patients in both treatment groups combined in the training set n Logistic model including treatment, selected main


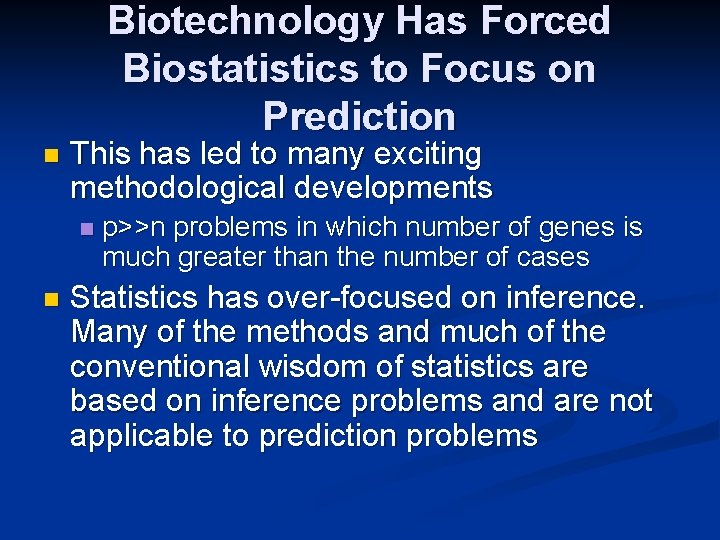
Biotechnology Has Forced Biostatistics to Focus on Prediction n This has led to many exciting methodological developments n n p>>n problems in which number of genes is much greater than the number of cases Statistics has over-focused on inference. Many of the methods and much of the conventional wisdom of statistics are based on inference problems and are not applicable to prediction problems

Prediction Based Clinical Trials n New methods for determining from RCTs which patients, if any, benefit from new treatments can be evaluated directly using the actual RCT data in a manner that separates model development from model evaluation, rather than basing treatment recommendations on the results of a single hypothesis test.

Prediction Based Clinical Trials n Using cross-validation we can evaluate new methods for analysis of clinical trials in terms of their intended use which is informing therapeutic decision making

Acknowledgements n Boris Freidlin n Yingdong Zhao n Wenyu Jiang n Aboubakar Maitournam
 Medicine
Medicine Phs human subjects and clinical trials information
Phs human subjects and clinical trials information Difference between inspection and audit
Difference between inspection and audit Nida clinical trials network
Nida clinical trials network Site initiation visit agenda
Site initiation visit agenda Clinical trial api
Clinical trial api Role of statistician in clinical trials
Role of statistician in clinical trials Andrew nunn
Andrew nunn Randomization in statistics
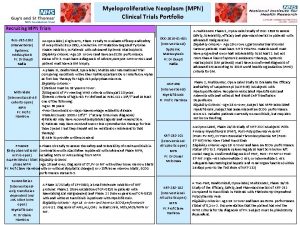
Randomization in statistics Mpn clinical trials
Mpn clinical trials Prs administrator
Prs administrator Clinical trials
Clinical trials Clinical trials quality by design
Clinical trials quality by design Mpn clinical trials
Mpn clinical trials Dhl atyrau
Dhl atyrau Clinical hysteria salem witch trials
Clinical hysteria salem witch trials Readyset ohsu
Readyset ohsu Clinical trials prs
Clinical trials prs Clinical trials.gov login
Clinical trials.gov login Ivr iwr studies
Ivr iwr studies York clinical trials unit
York clinical trials unit Principle of genomic equivalence
Principle of genomic equivalence Genomic england
Genomic england Genomic england
Genomic england Genomic england
Genomic england Genomic instability
Genomic instability Genomic
Genomic Genomic imprinting definition
Genomic imprinting definition Genomic signal processing
Genomic signal processing Comparative genomic hybridization animation
Comparative genomic hybridization animation Genomic equivalence definition
Genomic equivalence definition Looking for richard analysis
Looking for richard analysis Hexiangnan
Hexiangnan Personalized mobile search engine ieee paper
Personalized mobile search engine ieee paper Pentaho business analytics
Pentaho business analytics Personalized patient education
Personalized patient education Contextual bandits for personalized recommendation
Contextual bandits for personalized recommendation Personalized recommendations
Personalized recommendations Personalized navigation
Personalized navigation Personal statement first draft
Personal statement first draft All to one reduction
All to one reduction Trait approach leadership
Trait approach leadership Social stories questionnaire
Social stories questionnaire Personalized learning learner profile
Personalized learning learner profile Oyunlarm
Oyunlarm Institute for personalized learning
Institute for personalized learning Do we our life done
Do we our life done Design and analysis of cross-over trials
Design and analysis of cross-over trials Preventive and predictive maintenance of hydro power plant
Preventive and predictive maintenance of hydro power plant Concurrent criterion
Concurrent criterion Predictive analytics risk adjustment healthcare examples
Predictive analytics risk adjustment healthcare examples Health care risk adjustment and predictive modeling
Health care risk adjustment and predictive modeling Discovery education salem witch trials
Discovery education salem witch trials National geographic salem witch trials
National geographic salem witch trials Malta football trials
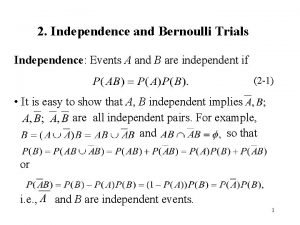
Malta football trials Repeated bernoulli trials
Repeated bernoulli trials Hercules hero's journey
Hercules hero's journey Future search trials
Future search trials Drug development timeline
Drug development timeline What caused the salem witch trial hysteria of 1692 dbq
What caused the salem witch trial hysteria of 1692 dbq Repeated bernoulli trials
Repeated bernoulli trials