Perl Introduction Why Perl Widely used scripting language




















































![Arrays my @fibonacci = (0, 1, 1, 2); print @fibonacci; print $fibonacci[3]; $fibonacci[4] = Arrays my @fibonacci = (0, 1, 1, 2); print @fibonacci; print $fibonacci[3]; $fibonacci[4] =](https://slidetodoc.com/presentation_image_h/8f183c591dc9abd851178baf6c986137/image-68.jpg)







![my @words = split(" ", "Hello World"); $words[0] = "Hello" $words[1] = "World" my @words = split(" ", "Hello World"); $words[0] = "Hello" $words[1] = "World"](https://slidetodoc.com/presentation_image_h/8f183c591dc9abd851178baf6c986137/image-87.jpg)
![More loops my @plant. List = ("rice", "potato", "tomato"); print $plant. List[0]; print $plant. More loops my @plant. List = ("rice", "potato", "tomato"); print $plant. List[0]; print $plant.](https://slidetodoc.com/presentation_image_h/8f183c591dc9abd851178baf6c986137/image-88.jpg)
































- Slides: 125

Perl Introduction

Why Perl? • • • Widely used scripting language Powerful text manipulation capabilities Relatively easy to use Has a wide range of libraries available Fast Good support for file and process operations

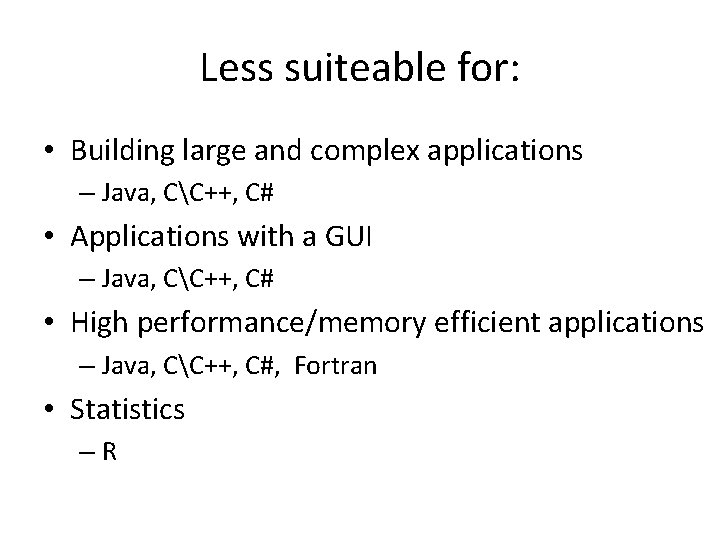
Less suiteable for: • Building large and complex applications – Java, CC++, C# • Applications with a GUI – Java, CC++, C# • High performance/memory efficient applications – Java, CC++, C#, Fortran • Statistics –R

Learning to script Knowledge + Skills

Exercise Determine the percentage GC-content of the human chromosome 22

open file read lines per line: skip if header line count Cs and Gs count all nucleotides report percentage Cs and Gs

Hello World

Hello World…. Simple line of Perl code: print "Hello World"; Run from a terminal: perl -e 'print "Hello World"; ' Now try this and notice the difference: perl -e 'print "Hello Worldn"; '

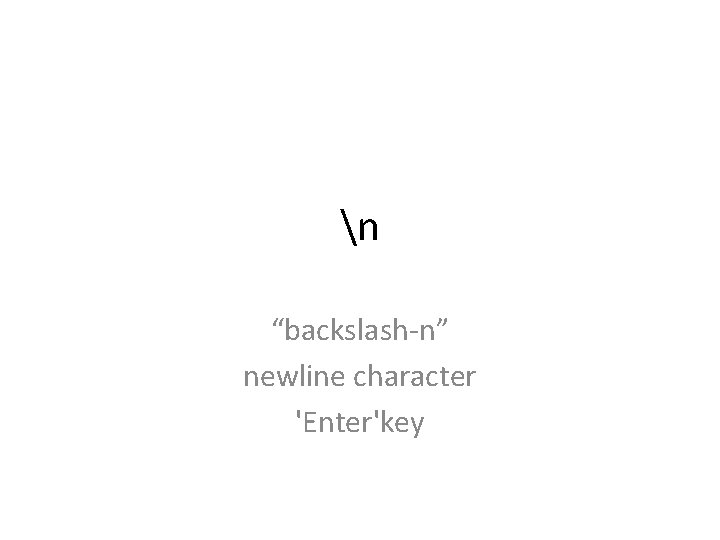
n “backslash-n” newline character 'Enter'key

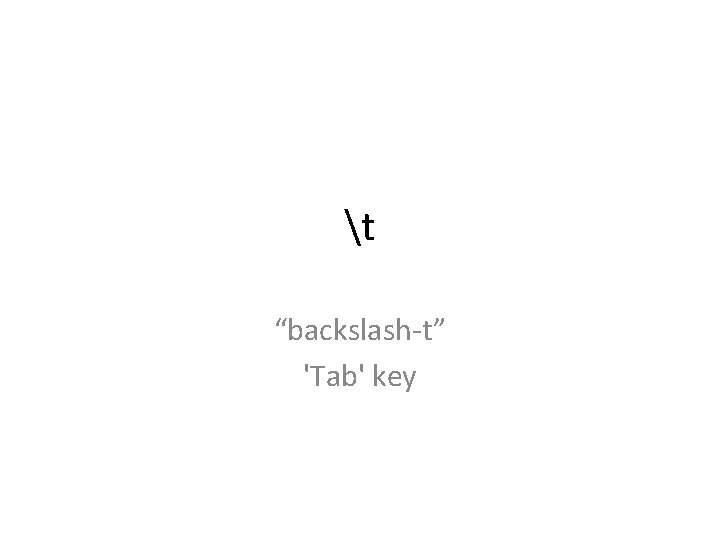
t “backslash-t” 'Tab' key

Hello World (cont) To create a text file with this line of Perl code: echo 'print "Hello Worldn"; ' > Hello. World. pl perl Hello. World. pl In the terminal window, type kate Hello. World. pl and then hit the enter key. Now you can edit the Perl code.

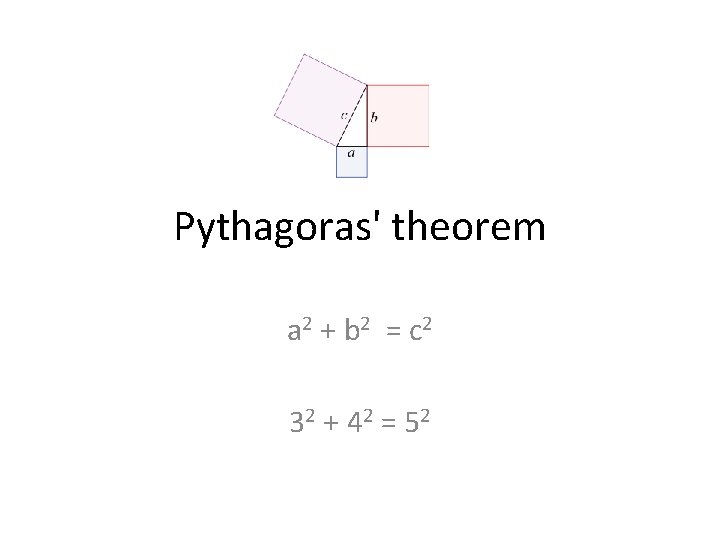
Pythagoras' theorem a 2 + b 2 = c 2 32 + 42 = 52

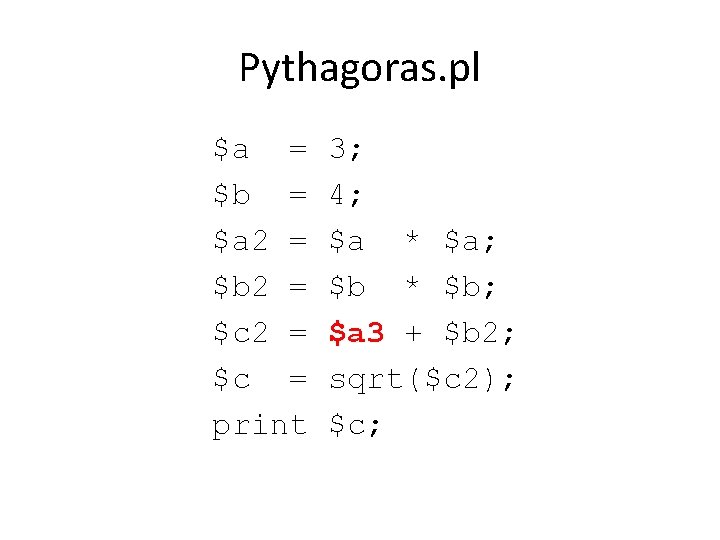
Pythagoras. pl $a = $b = $a 2 = $b 2 = $c = print 3; 4; $a * $a; $b * $b; $a 2 + $b 2; sqrt($c 2); $c;

$a a single value or scalar variable starts with a $ followed by its name

Pythagoras. pl $a = $b = $a 2 = $b 2 = $c = print 3; 4; $a * $a; $b * $b; $a 2 + $b 2; sqrt($c 2); $c;

5

Perl scripts Add these lines at the top of each Perl script: #!/usr/bin/perl # author: # description: use strict; use warnings;

perl Pythagoras. pl Global symbol "$a 2" requires explicit package name at Pythagoras. pl line 8. Global symbol "$b 2" requires explicit package name at Pythagoras. pl line 9. Global symbol "$c 2" requires explicit package name at Pythagoras. pl line 10. Global symbol "$a 2" requires explicit package name at Pythagoras. pl line 10. Global symbol "$b 2" requires explicit package name at Pythagoras. pl line 10. Global symbol "$c" requires explicit package name at Pythagoras. pl line 11. Global symbol "$c 2" requires explicit package name at Pythagoras. pl line 11. Global symbol "$c" requires explicit package name at Pythagoras. pl line 12. Execution of Pythagoras. pl aborted due to compilation errors.

Pythagoras. pl $a = $b = $a 2 = $b 2 = $c = print 3; 4; $a * $a; $b * $b; $a 2 + $b 2; sqrt($c 2); $c;

Pythagoras. pl my $a = 3; my $b = 4; my $a 2 = $a * $a; my $b 2 = $b * $b; my $c 2 = $a 2 + $b 2; my $c = sqrt($c 2); print $c;

my The first time a variable appears in the script, it should be claimed using ‘my’. Only the first time. . .

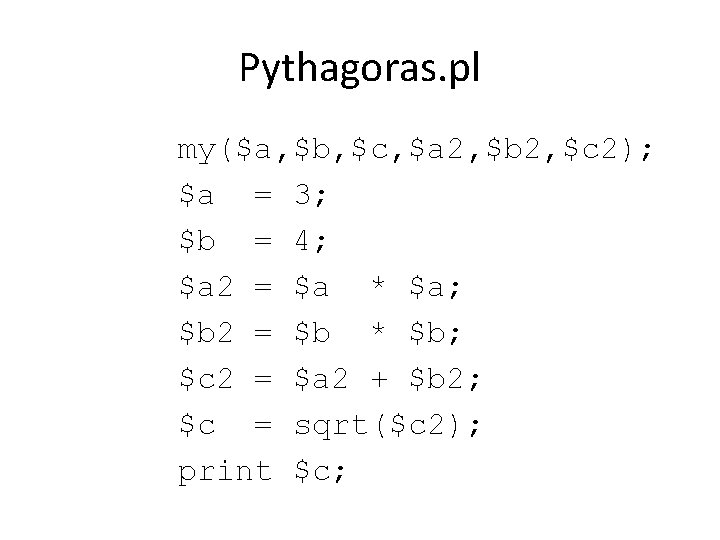
Pythagoras. pl my($a, $b, $c, $a 2, $b 2, $c 2); $a = 3; $b = 4; $a 2 = $a * $a; $b 2 = $b * $b; $c 2 = $a 2 + $b 2; $c = sqrt($c 2); print $c;

Pythagoras. pl $a = $b = $a 2 = $b 2 = $c = print 3; 4; $a * $a; $b * $b; $a 3 + $b 2; sqrt($c 2); $c;

4

Pythagoras. pl $a = $b = $a 2 = $b 2 = $c = print 3; 4; $a * $a; $b * $b; $a 3 + $b 2; sqrt($c 2); $c;

Pythagoras. pl my $a = 3; my $b = 4; my $a 2 = $a * $a; my $b 2 = $b * $b; my $c 2 = $a 3 + $b 2; my $c = sqrt($c 2); print $c;

perl Pythagoras. pl Global symbol "$a 3" requires explicit package name at Pythagoras. pl line 10. Execution of Pythagoras. pl aborted due to compilation errors.

Text or number Variables can contain text (strings) or numbers my $var 1 = 1; my $var 2 = "2"; my $var 3 = "three"; Try these four statements: print $var 1 + $var 2; print $var 2 + $var 3; print $var 1. $var 2; print $var 2. $var 3;

Text or number Variables can contain text (strings) or numbers my $var 1 = 1; my $var 2 = "2"; my $var 3 = "three"; Try these four statements: print $var 1 + $var 2; print $var 2 + $var 3; print $var 1. $var 2; print $var 2. $var 3; => => 3 2 12 2 three

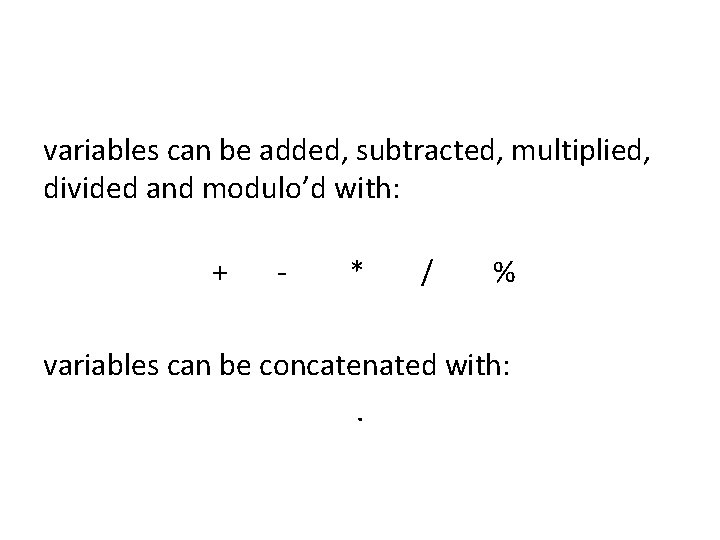
variables can be added, subtracted, multiplied, divided and modulo’d with: + - * / % variables can be concatenated with: .

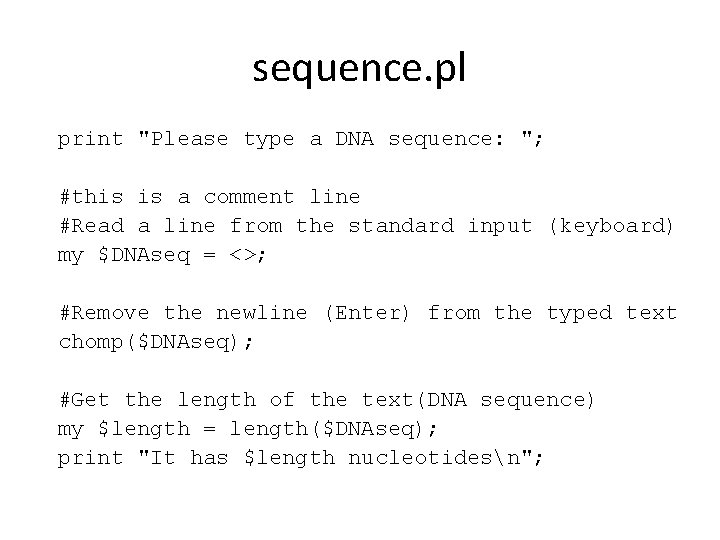
sequence. pl print "Please type a DNA sequence: "; #this is a comment line #Read a line from the standard input (keyboard) my $DNAseq = <STDIN>; #Remove the newline (Enter) from the typed text chomp($DNAseq); #Get the length of the text(DNA sequence) my $length = length($DNAseq); print "It has $length nucleotidesn";

Program flow is top - down sequence. pl print "Please type a DNA sequence: "; #this is a comment line #Read a line from the standard input (keyboard) my $DNAseq = <STDIN>; #Remove the newline (Enter) from the typed text chomp($DNAseq); #Get the length of the text(DNA sequence) my $length = length($DNAseq); print "It has $length nucleotidesn";

<STDIN> read characters that are typed on the keyboard. Stop after the Enter key is pressed

<> same, STDIN is the default and can be left out. This is a recurring and confusing theme in Perl. . .

sequence. pl print "Please type a DNA sequence: "; #this is a comment line #Read a line from the standard input (keyboard) my $DNAseq = <>; #Remove the newline (Enter) from the typed text chomp($DNAseq); #Get the length of the text(DNA sequence) my $length = length($DNAseq); print "It has $length nucleotidesn";

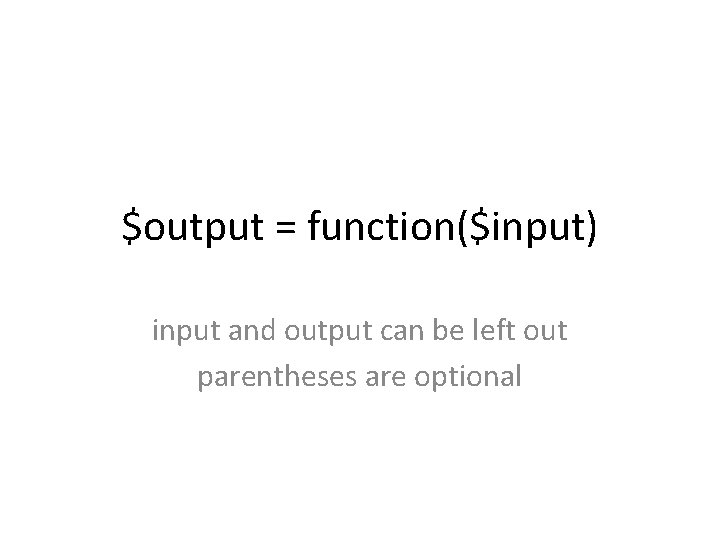
$output = function($input) input and output can be left out parentheses are optional

$coffee = function($beans, $water)

sequence 2. pl print "Please type a DNA sequence: "; my $DNAseq = <>; chomp($DNAseq); #Get the first three characters of $DNAseq my $first 3 bases = substr($DNAseq, 0, 3); print "The first 3 bases: $first 3 basesn";

$frag = substr($text, $start, $num) Extract a fragment of string $text starting at $start and with $num characters. The first letter is at position 0!

perldoc -f substr EXPR, OFFSET, LENGTH, REPLACEMENT substr EXPR, OFFSET, LENGTH substr EXPR, OFFSET Extracts a substring out of EXPR and returns it. First character is at offset 0, . . .

print perldoc -f print FILEHANDLE LIST print Prints a string or a list of strings. If you leave out the FILEHANDLE, STDOUT is the destination: your terminal window.

print In Perl items in a list are separated by commas print "Hello World", "n"; Is the same as: print "Hello Worldn";

sequence 3. pl print "Please type a DNA sequence: "; my $DNAseq = <>; chomp($DNAseq); #Get the second codon of $DNAseq my $codon 2 = substr($DNAseq, 3, 3); print "The second codon: $codon 2n";

if, else, unless

sequence 4. pl print "Please type a DNA sequence: "; my $DNAseq = <>; chomp($DNAseq); #Get the first three characters of $DNAseq my $codon = substr($DNAseq, 0, 3); if($codon eq "ATG") { print "Found a start codonn"; }

Conditional execution if ( condition ) { do something } else { do something else }

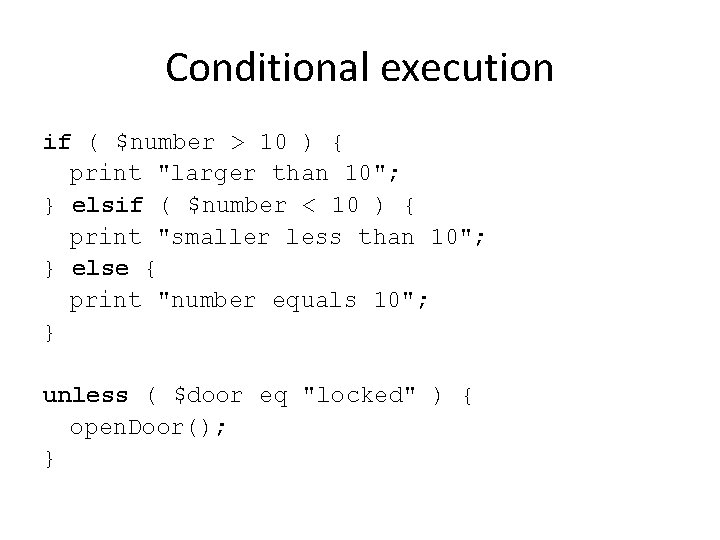
Conditional execution if ( $number > 10 ) { print "larger than 10"; } elsif ( $number < 10 ) { print "smaller less than 10"; } else { print "number equals 10"; } unless ( $door eq "locked" ) { open. Door(); }

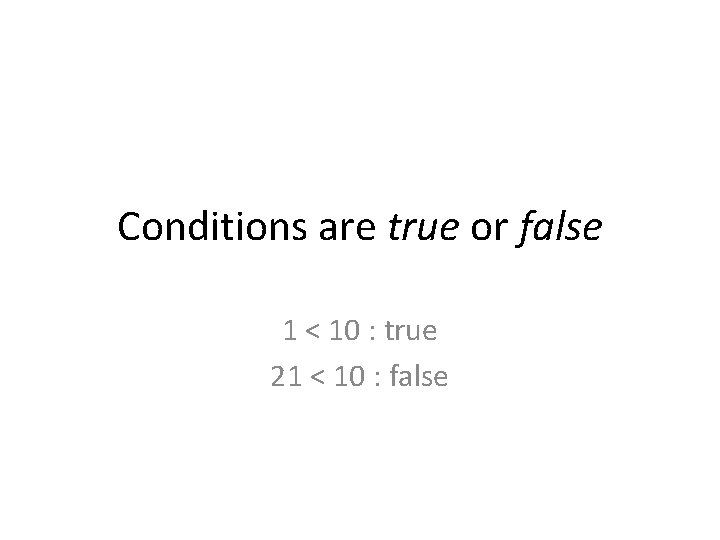
Conditions are true or false 1 < 10 : true 21 < 10 : false

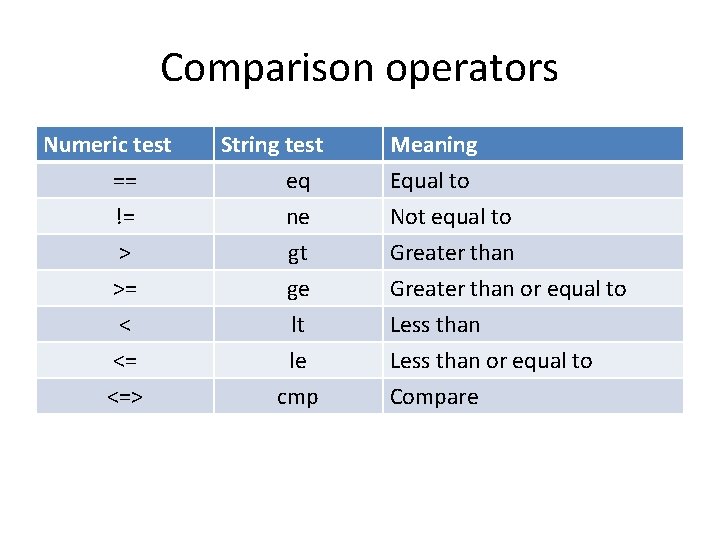
Comparison operators Numeric test == != > >= < <= <=> String test eq ne gt ge lt le cmp Meaning Equal to Not equal to Greater than or equal to Less than or equal to Compare

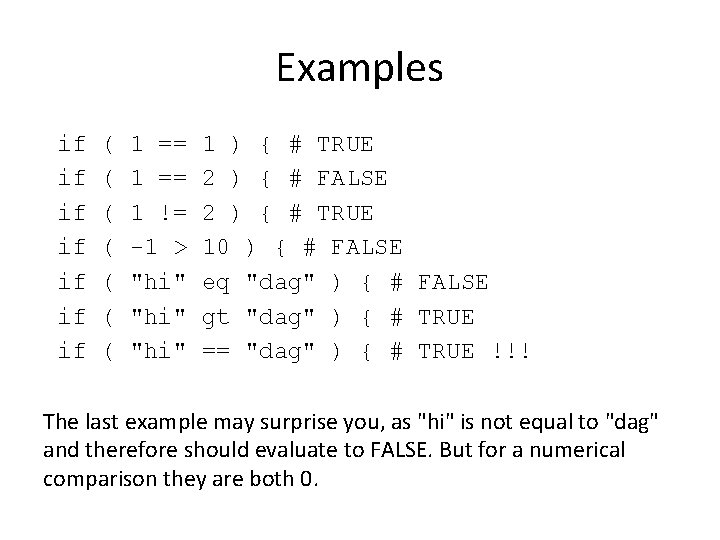
Examples if if ( ( ( ( 1 == 1 != -1 > "hi" 1 ) { # TRUE 2 ) { # FALSE 2 ) { # TRUE 10 ) { # FALSE eq "dag" ) { # FALSE gt "dag" ) { # TRUE == "dag" ) { # TRUE !!! The last example may surprise you, as "hi" is not equal to "dag" and therefore should evaluate to FALSE. But for a numerical comparison they are both 0.

numbers as conditions 0 : false all other numbers : true

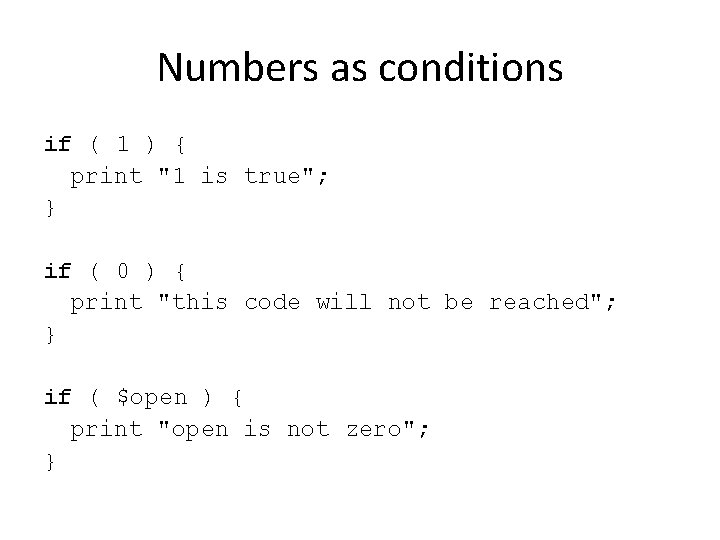
Numbers as conditions if ( 1 ) { print "1 is true"; } if ( 0 ) { print "this code will not be reached"; } if ( $open ) { print "open is not zero"; }

repetition

sequence 5. pl print "Please type a DNA sequence: "; my $DNAseq = <>; chomp($DNAseq); #Get all codons of $DNAseq my $position = 0 while($position < length($DNAseq)) { my $codon = substr($DNAseq, $position, 3); print "The next codon: $codonn"; $position = $position + 3; }

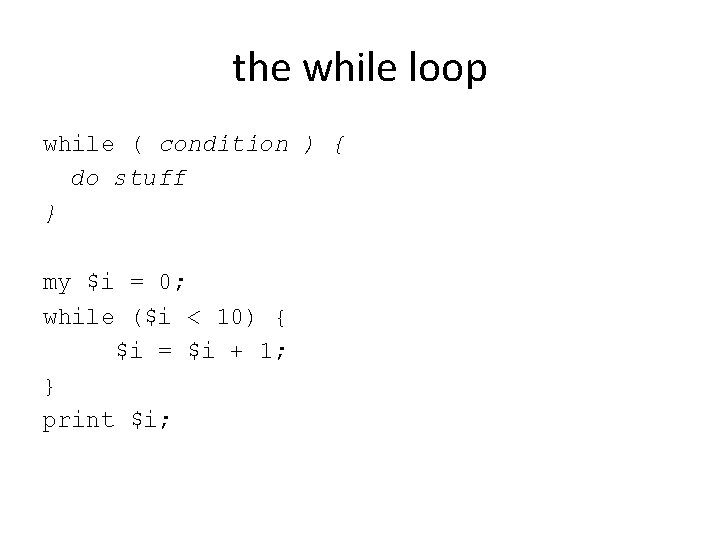
the while loop while ( condition ) { do stuff } my $i = 0; while ($i < 10) { $i = $i + 1; } print $i;

$i = $i + 1 First the part to the right of the assignment operator ‘=‘ is calculated, then the result is moved to the left.

$i += 1 Same result as previous slide.

$i++ Same as result previous slide, increments $i with 1.

++$i Same as previous, but compare: print $i++; print ++$i;

Exercise: Fibonacci numbers Write a script that calculates and prints all Fibonacci numbers below one thousand. 0, 1, 1, 2, 3, 5, 8, 13, 21, 34, 55, 89, 144, etc. Fn = Fn-1+ Fn-2 F 0 = 0, F 1 = 1

sequence 5. pl print "Please type a DNA sequence: "; my $DNAseq = <>; chomp($DNAseq); #Copy the sequence to a new variable my $as. DNAseq = $DNAseq; #'translate' a->t, c->g, g->c, t->a $as. DNAseq =~ tr/acgt/tgca/; print "Complementary strand: n$as. DNAseqn";

$as. DNAseq =~ tr/acgt/tgca/; =~ is a binding operator and means: perform the following action on this variable. The operation tr/// translates each character from the first set of characters into the corresponding character in the second set: acgt |||| tgca

Counting tr/// can also be used to count characters. If the second part is left empty, no translation takes place. $number. Of. Ns = ($DNASeq =~ tr/N//);

'automatic' typing using a pipe "|": echo ggatcc | perl sequence 5. pl or redirect using "<": perl sequence 5. pl < sequence. txt

Exercise 1. Create a program that reads a DNA sequence from the keyboard, and reports the sequence length and the G/C content of the sequence (as a fraction)

perltidy program that properly formats your perl script Indentation, spaces, etc. perltidy yourscript. pl Result is in: yourscript. pl. tdy

0 1 2 @months 3 a list variable or array starts with an @ followed by its name
![Arrays my fibonacci 0 1 1 2 print fibonacci print fibonacci3 fibonacci4 Arrays my @fibonacci = (0, 1, 1, 2); print @fibonacci; print $fibonacci[3]; $fibonacci[4] =](https://slidetodoc.com/presentation_image_h/8f183c591dc9abd851178baf6c986137/image-68.jpg)
Arrays my @fibonacci = (0, 1, 1, 2); print @fibonacci; print $fibonacci[3]; $fibonacci[4] = 3; $fibonacci[5] = 5; $fibonacci[6] = 8;

@fibonacci 0 0 1 1 2 1 3 2

Arrays my @hw = ("Hello ", "World", "n"); print @hw; my @months = ( "January", "February", "March");

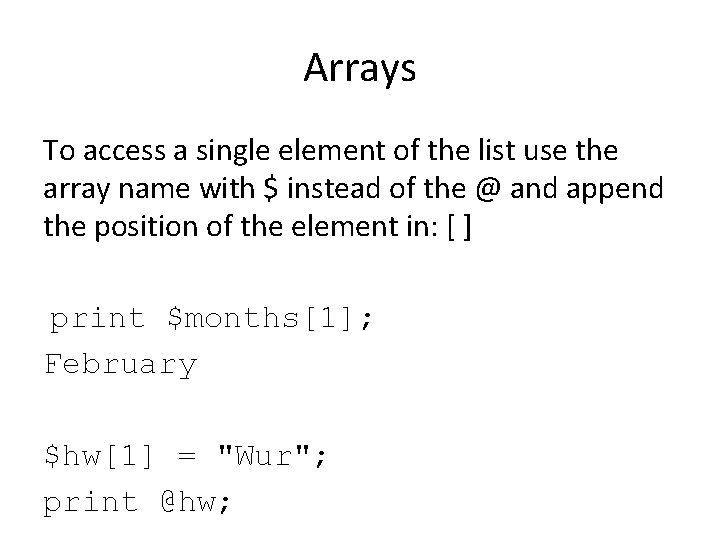
Arrays To access a single element of the list use the array name with $ instead of the @ and append the position of the element in: [ ] print $months[1]; February $hw[1] = "Wur"; print @hw;

Arrays To find the index of the last element in the list: print $#months; 2 To find the number of elements in an array: print $#months + 1; or: print scalar(@months);

Arrays Note: like many programming languages, the index of the first item in an array is not 1, but 0! Note: $months ≠ $months[0] !!!

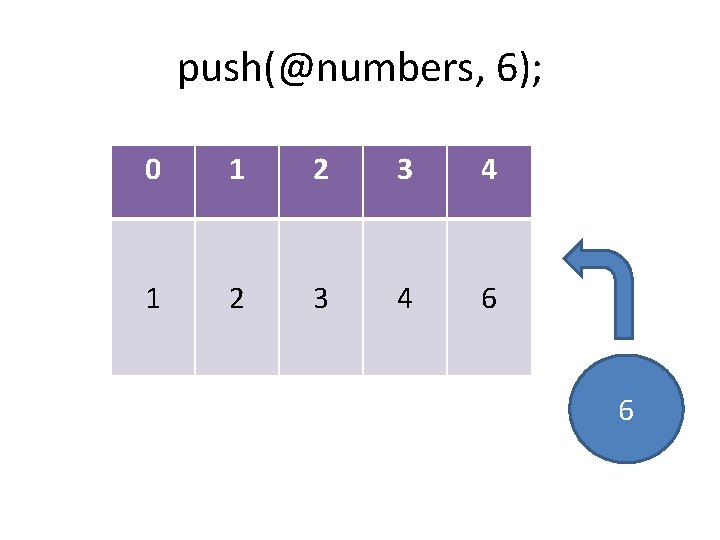
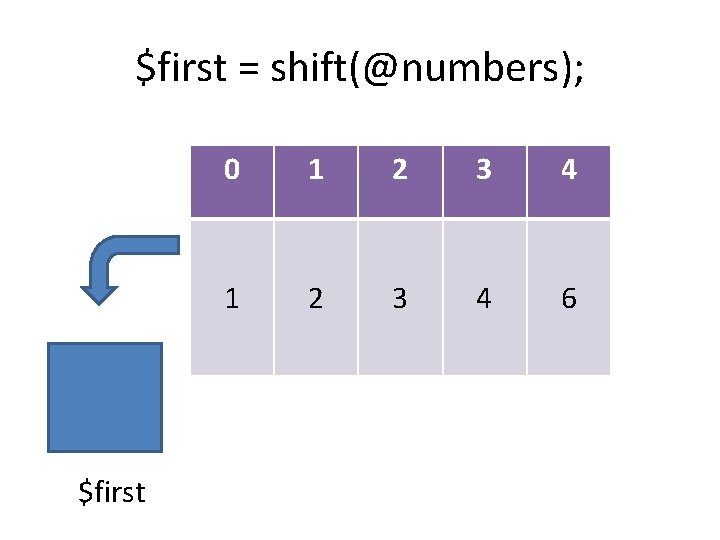
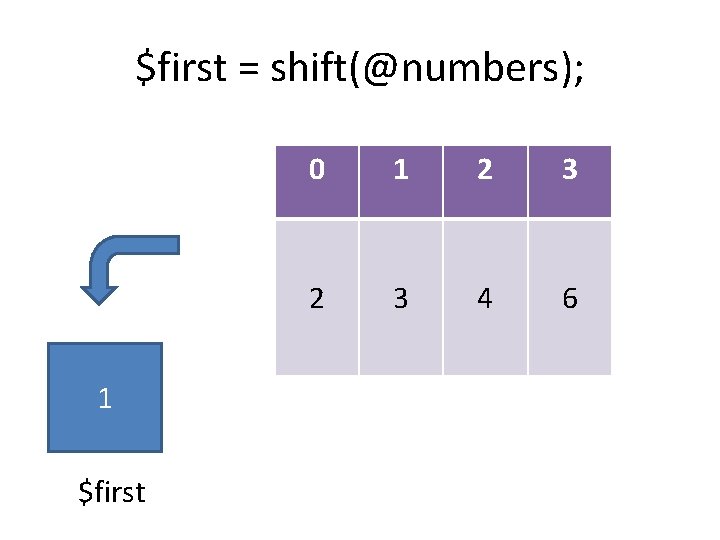
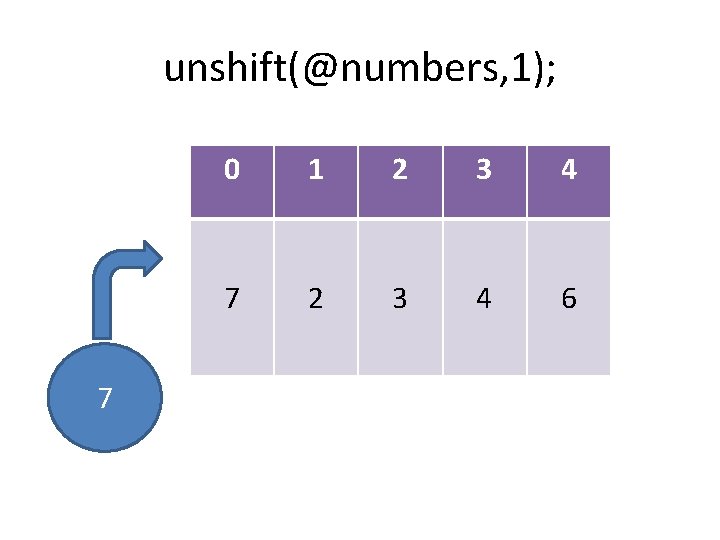
Growing and shrinking arrays push: pop: shift: unshift: splice: add an item to the end of the list remove an item from the start of the list add an item to the start of the list insert/remove one or more items @out = splice(@array, start, length, @in);

@numbers index 0 1 2 3 4 value 1 2 3 4 5

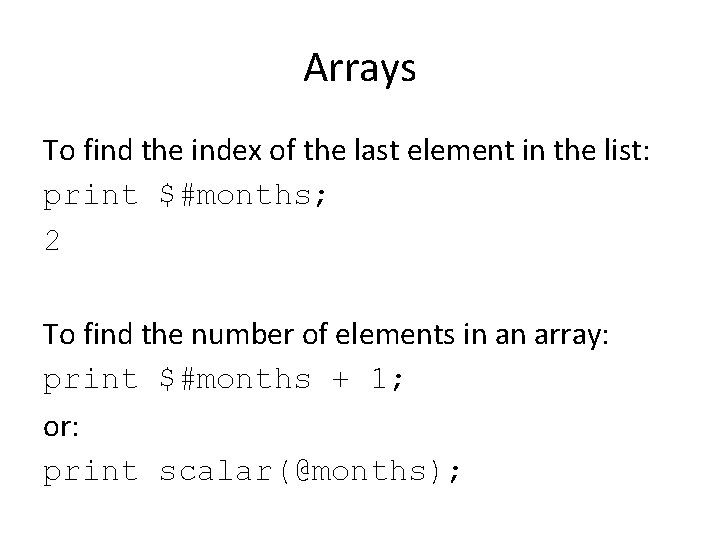
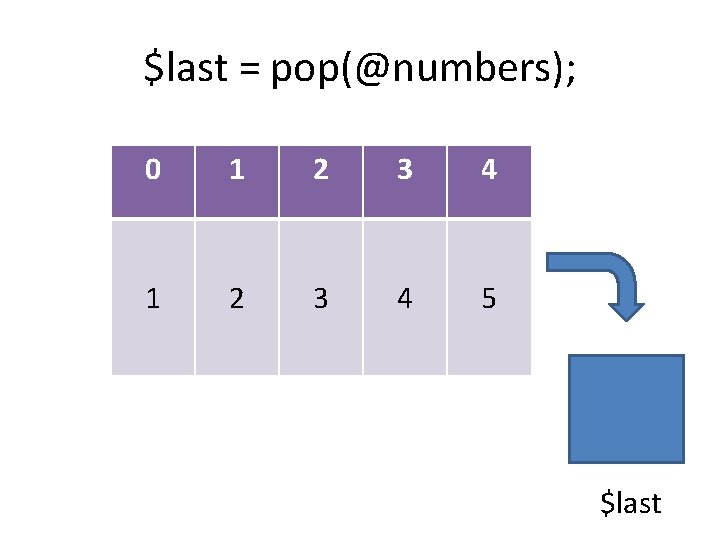
$last = pop(@numbers); 0 1 2 3 4 5 $last

$last = pop(@numbers); 0 1 2 3 4 5 $last

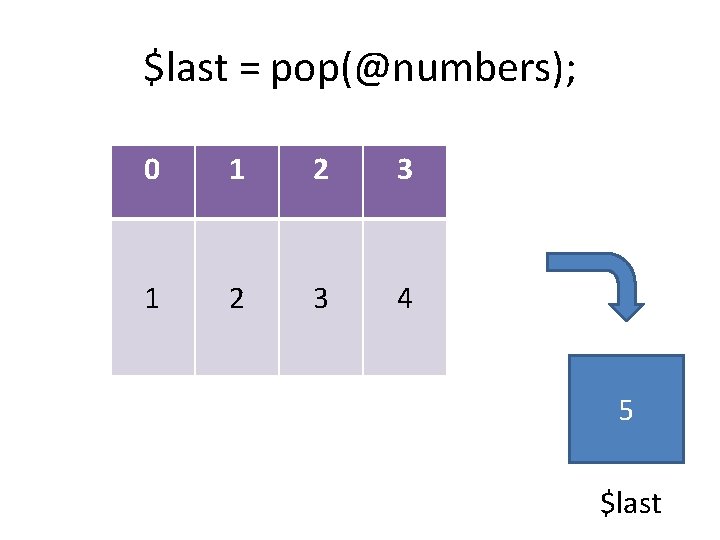
push(@numbers, 6); 0 1 2 3 4 6

push(@numbers, 6); 0 1 2 3 4 6 6

$first = shift(@numbers); $first 0 1 2 3 4 6

$first = shift(@numbers); 1 $first 0 1 2 3 4 6

unshift(@numbers, 7); 7 0 1 2 3 4 6

unshift(@numbers, 1); 7 0 1 2 3 4 7 2 3 4 6

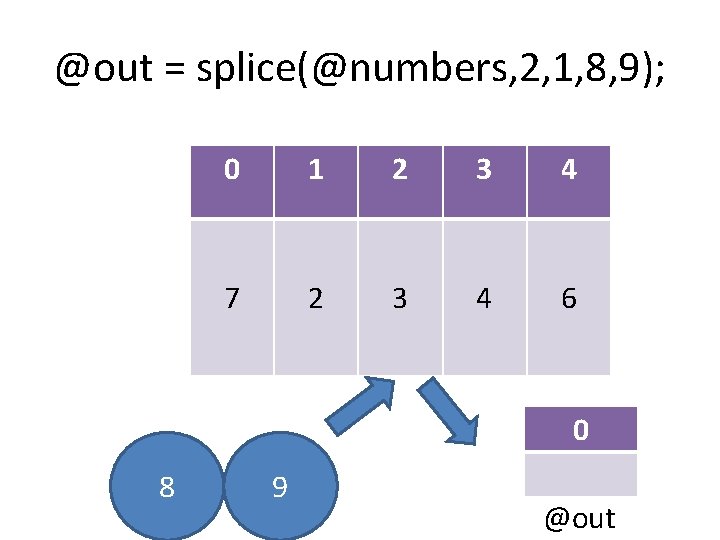
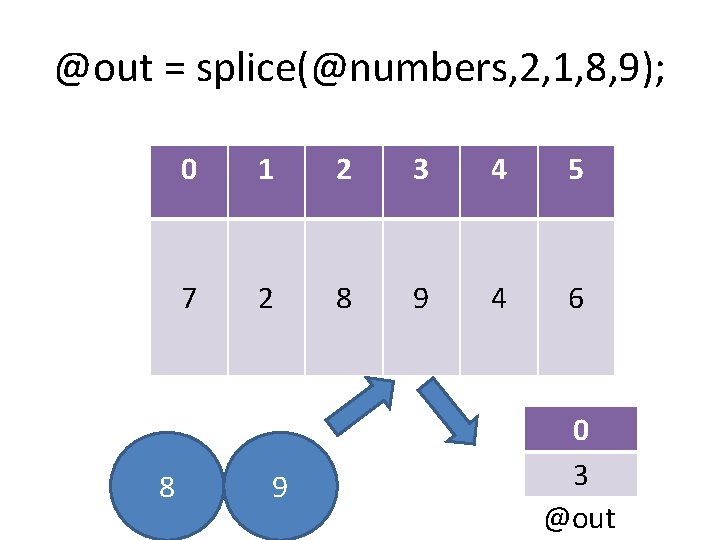
@out = splice(@numbers, 2, 1, 8, 9); 0 1 2 3 4 7 2 3 4 6 0 8 9 @out

@out = splice(@numbers, 2, 1, 8, 9); 8 0 1 2 3 4 5 7 2 8 9 4 6 9 0 3 @out

my ($x, $y, $z) = @coordinates;
![my words split Hello World words0 Hello words1 World my @words = split(" ", "Hello World"); $words[0] = "Hello" $words[1] = "World"](https://slidetodoc.com/presentation_image_h/8f183c591dc9abd851178baf6c986137/image-87.jpg)
my @words = split(" ", "Hello World"); $words[0] = "Hello" $words[1] = "World"
![More loops my plant List rice potato tomato print plant List0 print plant More loops my @plant. List = ("rice", "potato", "tomato"); print $plant. List[0]; print $plant.](https://slidetodoc.com/presentation_image_h/8f183c591dc9abd851178baf6c986137/image-88.jpg)
More loops my @plant. List = ("rice", "potato", "tomato"); print $plant. List[0]; print $plant. List[1]; Print $plant. List[2]; Or: foreach my $plant (@plant. List) { print $plant; }

Loops foreach variable ( list ) { do something with the variable } foreach my $i ( @lotto_numbers ) { print $i; } foreach my $i ( 1. . 10, 20, 30 ) { print $i; }

Loops for variable ( list ) { do something with the variable } for my $i ( 1, 2, 3, 4, 5 ) { print $i; } for my $i ( 1. . 10, 20, 30 ) { print $i; }

Loops while ( condition ) { do something } my $i = 0; while ($i < 10) { print "$i < 10n"; $i++; }

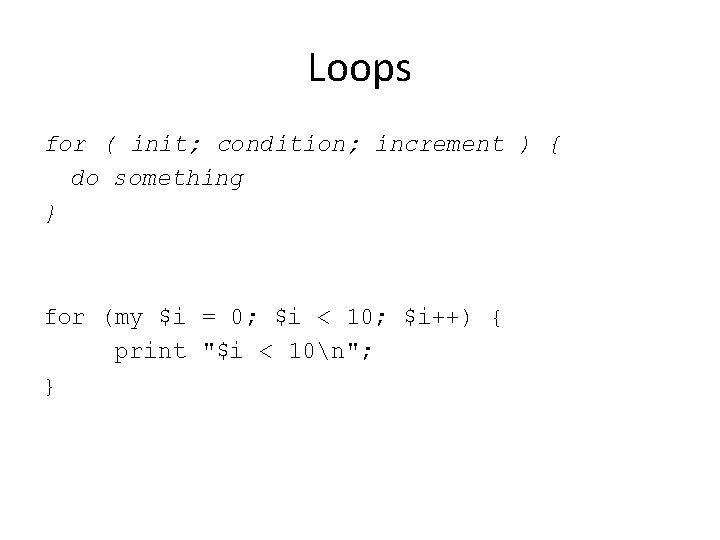
Loops for ( init; condition; increment ) { do something } for (my $i = 0; $i < 10; $i++) { print "$i < 10n"; }

Loops my $i = 0; while ($i < 10) { print "$i < 10n"; $i++; } for (my $i = 0; $i < 10; $i++) { print "$i < 10n"; }

Exercise Write a script that reverses a DNA sequence use an array Hint: Splitting on an empty string "" splits after every character. @sequence = split("", $sequence);

0 1 2 3 Name Box Crick 3 Franklin 1 Watson 0 Wilkins 2 %phonebook a hash table variable starts with a % followed by its name

Hash tables Also called lookup tables, dictionaries or associative arrays key/value combinations: keys are text, values can be anything %month_days = ("January" => 31, "February" => 28, "March" => 31 );

Hash tables To access a value in the hash table, use the hash table name with $ instead of the % and append the key between { } $month_days{"February"} = 29; print $month_days{"January"}; 31

Hash tables The 'keys' function returns an list with the keys of the hash table. There is also a 'values' function. @month_list = keys(%month_days); # ("January", "February", "March")

Hash tables my %latin_name=( "rice" => "Oryza sativa", "potato" => "Solanum tuberosum" ) foreach my $common_name (keys(%latin_name)){ print "$common_name: " ; print "$latin_name{$common_name}n"; } rice: Oryza sativa potato: Solanum tuberosum

Hash tables The keys have to be unique, the values do not. The order of elements in a hash table is not reliable, first in is not necessarily first out. You can use 'sort' to get the keys in an alphabetically ordered list: @sorted = sort(keys(%latin_name));

Exercise Create a hash table with codons as keys and the corresponding amino acids as the values Hint: search for the standard genetic code in the "genetic code" database at: http: //srs. bioinformatics. nl/ Use three lines for the first, second and third base and the line for the corresponding AA.

I/O: Input and Output reading and writing files

Reading and writing files open FASTA, "sequence. fa"; my $first. Line = <FASTA>; my $second. Line = <FASTA>; close FASTA;

Reading and writing files Files need to be opened before use

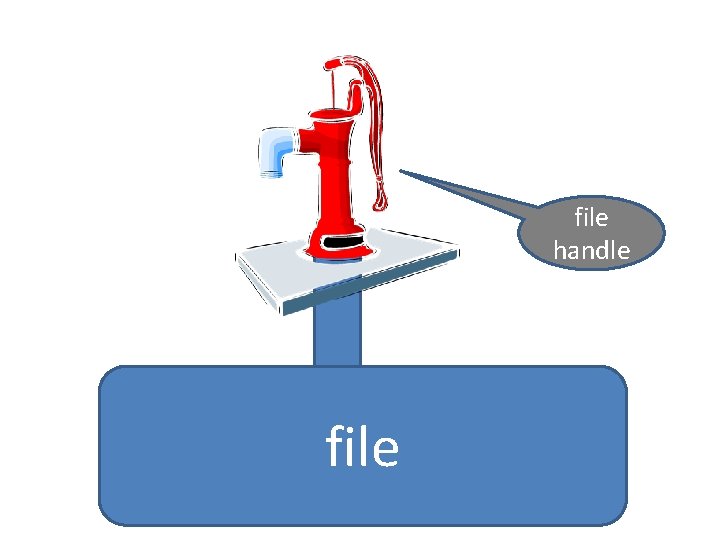
Reading and writing files Perl uses so-called “file handles” to attach to files for reading and writing

file handle file

Opening files General open File. Handle, "mode", "filename" Open for reading: open LOG, "<", "/var/log/messages"; open LOG, "/var/log/messages"; Open for writing: open WRT, ">", "newfile. txt"; Open for appending: open APP, ">>", "existingfile. txt";

Defensive programming my $fasta. Name = "sequence. fa"; open FASTA, $fasta. Name or die "cannot open $fasta. Namen";

Reading from a file reading from an open file via the filehandle: $first. Line = <FASTA>; $second. Line = <FASTA>; @other. Lines = <FASTA>;

<FASTA> Reads one line if the result goes into a scalar $line = <FASTA>; Reads all (remaining) lines if the result goes into an array @lines = <FASTA>; file handles 'remember' the position in the file

Standard in and standard out The keyboard and screen also have 'file' handles, remember STDIN and STDOUT read from the keyboard: $DNAseq = <STDIN>; write to the screen: print STDOUT "Hello Worldn";

Reading a file open FASTA, "sequence. fa" or die; my $sequence = ""; while (my $line = <FASTA>) { chomp($line); $sequence. = $line; } close FASTA; print $sequence, "n";

(my $line = <FASTA>) also is a condition true: line could be read false: EOF, end of file

Identical? while (my $line = print $line; } <FASTA>) { for my $line (<FASTA>) { print $line; }

Not completely Read line by line: while (my $line = <FASTA>) { print $line; } First read complete file into computer memory: for my $line (<FASTA>) { print $line; }

Writing to a file open RANDOM, ">", "Random. txt"; for(1. . 50) { my $random = rand(6); print RANDOM "$randomn"; } close RANDOM;

Writing to a file open RANDOM, ">", "Random. txt"; for(1. . 50) { my $rnd = rand(6); $rnd = sprintf("%dn", $rnd + 1); print RANDOM $rnd; } close RANDOM;

Closing the file close filehandle; close FASTA; A file is automatically closed if you (re)open a file using the same filehandle, or if the Perl script is finished.

Minimalistic Perl open FASTA, "sequence. fa" or die; my $sequence = ""; while (my $line = <FASTA>) { chomp($line); $sequence. = $line; } close FASTA; print $sequence, "n";

Minimalistic Perl open FASTA, "sequence. fa" or die; my $sequence = ""; while (<FASTA>) { chomp; $sequence. = $_; } close FASTA; print $sequence, "n";

$_ default scalar variable, if no other variable is given. But only in selected cases. . .

Minimalistic Perl open FASTA, "sequence. fa" or die; my $sequence = ""; while (<FASTA>) { chomp; $sequence. = $_; } close FASTA; print $sequence, "n";

Minimalistic Perl open FASTA, "sequence. fa" or die; my $sequence = ""; while ($_ = <FASTA>) { chomp($_); $sequence. = $_; } close FASTA; print $sequence, "n";

Exercises 2. Adapt the G/C script so multiple sequences in FASTA format are read from a file 3. Modify the script to process a file containing any number of sequences in EMBL format 4. Now let the program generate the reverse complement of the sequence(s), and report sequence length and G/C content

Exercises 5. Use the rand function of Perl to shuffle the nucleotides of the input sequence, while maintaining sequence composition; again report sequence length and G/C content