Peptoids that Mimic the Structure Function and Mechanism

- Slides: 23

Peptoids that Mimic the Structure, Function, and Mechanism of Helical Antimicrobial Peptides PNAS. 2008. Vol. 105(8). 2794 -2799

Antimicrobial Peptides Are natural peptides that defend the host organism against bacterial infection. n They typical contain both positively charged and hydrophobic residues. n Interact with the cellular membrane of the respective pathogens in a selective manner. n

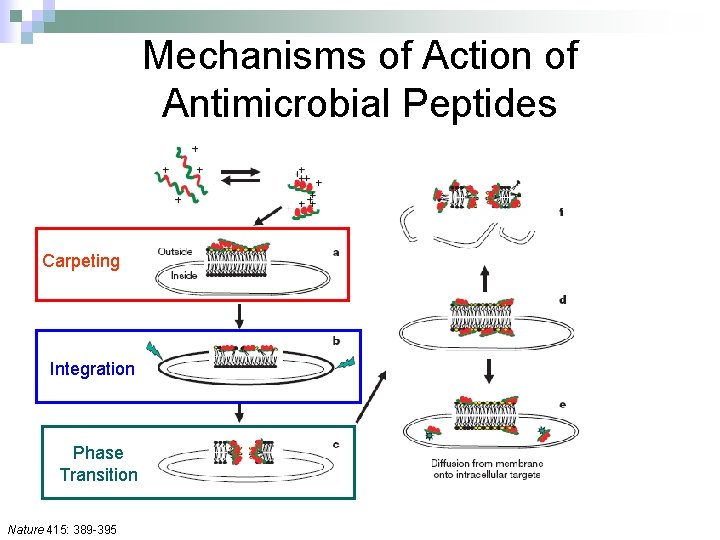
Mechanisms of Action of Antimicrobial Peptides Carpeting Integration Phase Transition Nature 415: 389 -395

Proteolysis of CAMPs n Cationic antimicrobial peptides are very susceptible to proteolytic degradation by bacterial and host proteases. Nature Reviews Microbiology 4, 529 -536 (July 2006)

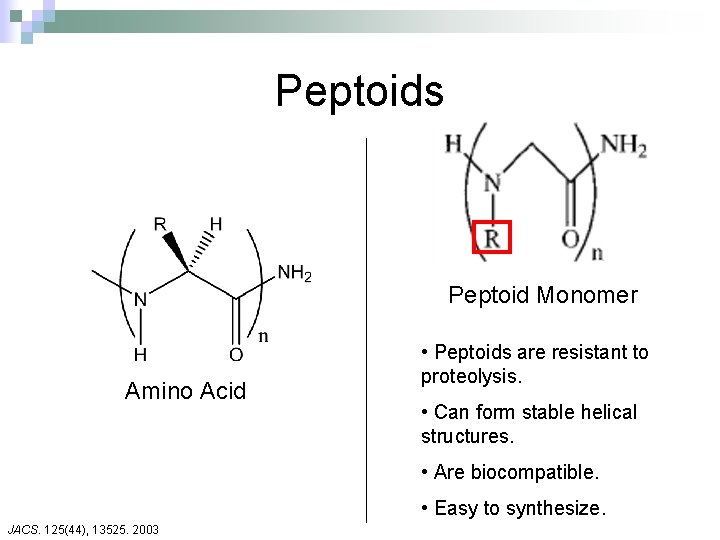
Peptoids Peptoid Monomer Amino Acid • Peptoids are resistant to proteolysis. • Can form stable helical structures. • Are biocompatible. • Easy to synthesize. JACS. 125(44), 13525. 2003

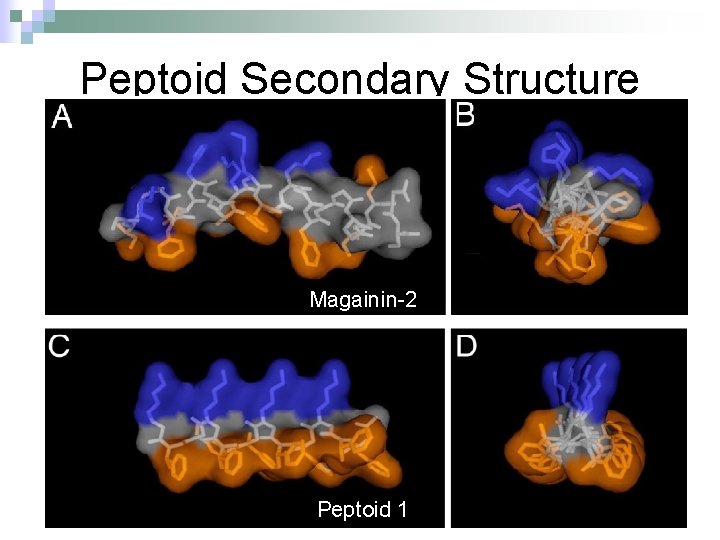
Peptoid Secondary Structure Magainin-2 Peptoid 1

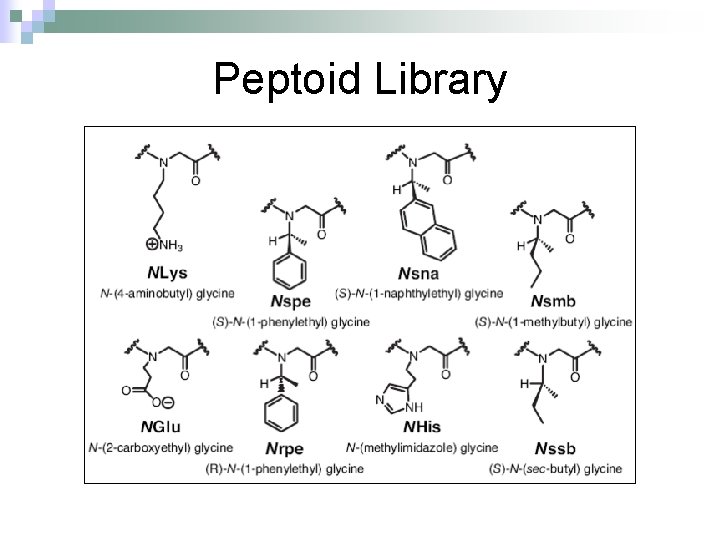
Peptoid Library

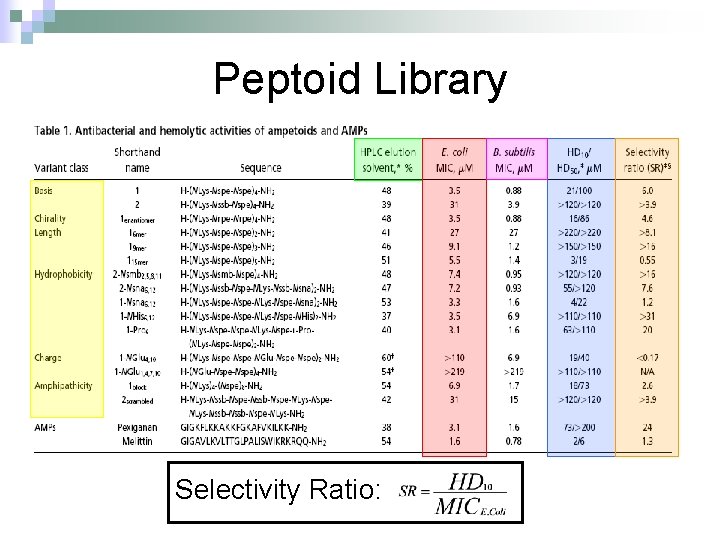
Peptoid Library Selectivity Ratio:

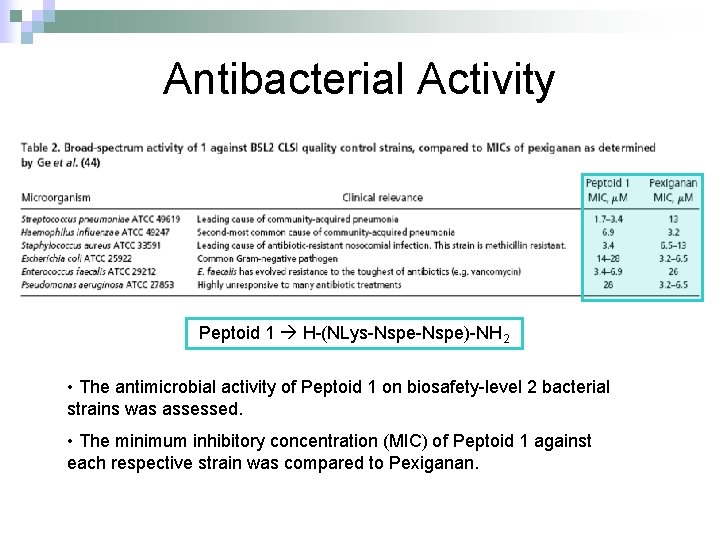
Antibacterial Activity Peptoid 1 H-(NLys-Nspe)-NH 2 • The antimicrobial activity of Peptoid 1 on biosafety-level 2 bacterial strains was assessed. • The minimum inhibitory concentration (MIC) of Peptoid 1 against each respective strain was compared to Pexiganan.

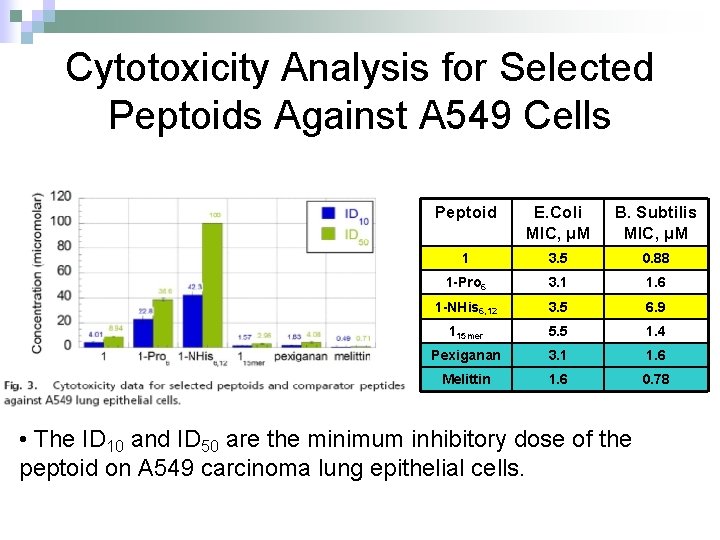
Cytotoxicity Analysis for Selected Peptoids Against A 549 Cells Peptoid E. Coli MIC, μM B. Subtilis MIC, μM 1 3. 5 0. 88 1 -Pro 6 3. 1 1. 6 1 -NHis 6, 12 3. 5 6. 9 115 mer 5. 5 1. 4 Pexiganan 3. 1 1. 6 Melittin 1. 6 0. 78 • The ID 10 and ID 50 are the minimum inhibitory dose of the peptoid on A 549 carcinoma lung epithelial cells.

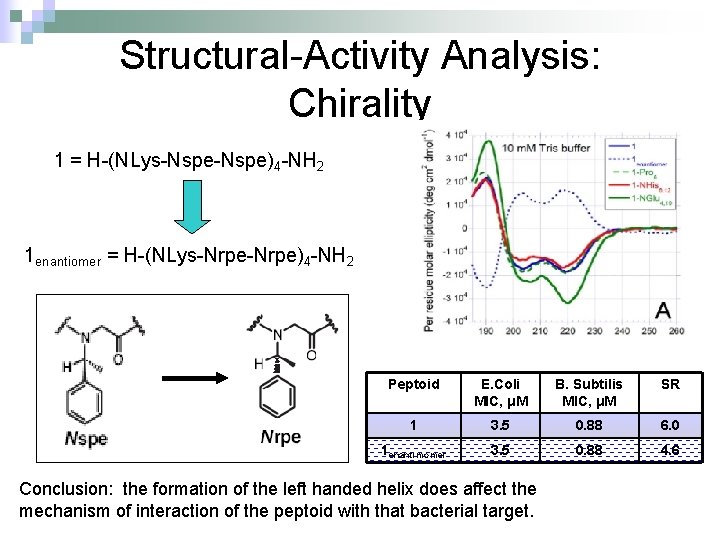
Structural-Activity Analysis: Chirality 1 = H-(NLys-Nspe)4 -NH 2 1 enantiomer = H-(NLys-Nrpe)4 -NH 2 Peptoid E. Coli MIC, μM B. Subtilis MIC, μM SR 1 3. 5 0. 88 6. 0 1 enantimomer 3. 5 0. 88 4. 6 Conclusion: the formation of the left handed helix does affect the mechanism of interaction of the peptoid with that bacterial target.

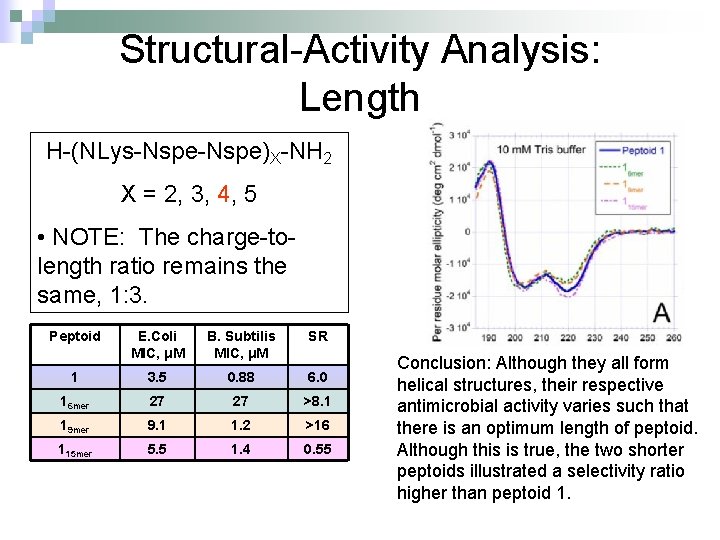
Structural-Activity Analysis: Length H-(NLys-Nspe)X-NH 2 X = 2, 3, 4, 5 • NOTE: The charge-tolength ratio remains the same, 1: 3. Peptoid E. Coli MIC, μM B. Subtilis MIC, μM SR 1 3. 5 0. 88 6. 0 16 mer 27 27 >8. 1 19 mer 9. 1 1. 2 >16 115 mer 5. 5 1. 4 0. 55 Conclusion: Although they all form helical structures, their respective antimicrobial activity varies such that there is an optimum length of peptoid. Although this is true, the two shorter peptoids illustrated a selectivity ratio higher than peptoid 1.

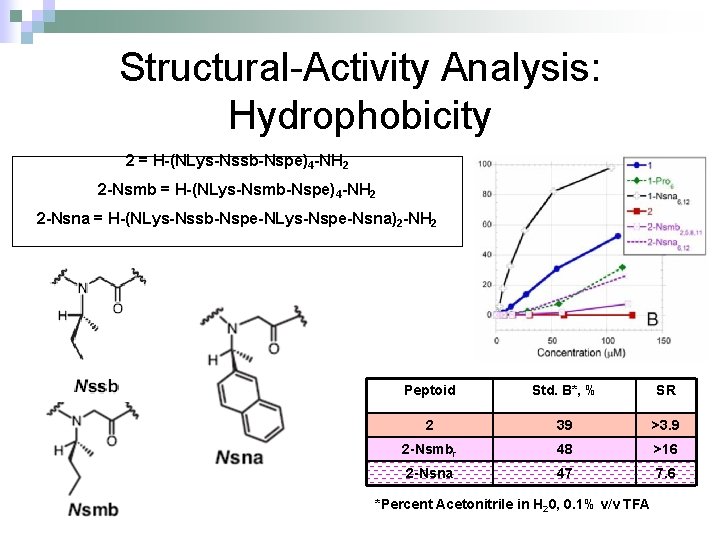
Structural-Activity Analysis: Hydrophobicity 2 = H-(NLys-Nssb-Nspe)4 -NH 2 2 -Nsmb = H-(NLys-Nsmb-Nspe)4 -NH 2 2 -Nsna = H-(NLys-Nssb-Nspe-NLys-Nspe-Nsna)2 -NH 2 Peptoid Std. B*, % SR 2 39 >3. 9 2 -Nsmbr 48 >16 2 -Nsna 47 7. 6 *Percent Acetonitrile in H 20, 0. 1% v/v TFA

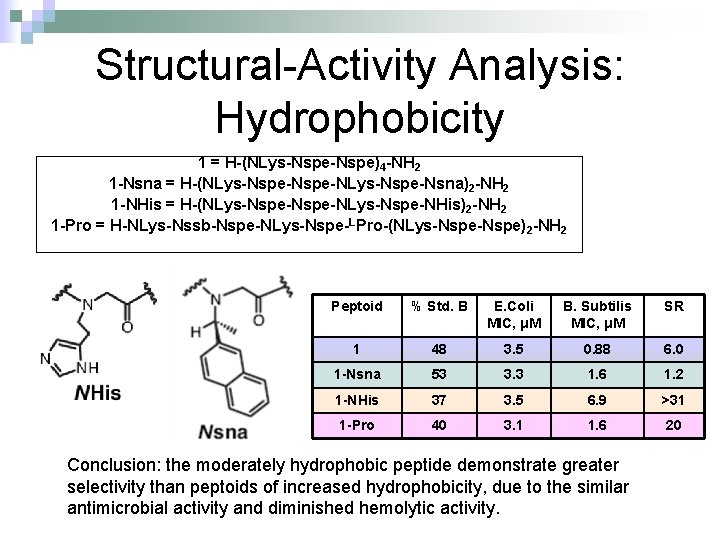
Structural-Activity Analysis: Hydrophobicity 1 = H-(NLys-Nspe)4 -NH 2 1 -Nsna = H-(NLys-Nspe-NLys-Nspe-Nsna)2 -NH 2 1 -NHis = H-(NLys-Nspe-NLys-Nspe-NHis)2 -NH 2 1 -Pro = H-NLys-Nssb-Nspe-NLys-Nspe-LPro-(NLys-Nspe)2 -NH 2 Peptoid % Std. B E. Coli MIC, μM B. Subtilis MIC, μM SR 1 48 3. 5 0. 88 6. 0 1 -Nsna 53 3. 3 1. 6 1. 2 1 -NHis 37 3. 5 6. 9 >31 1 -Pro 40 3. 1 1. 6 20 Conclusion: the moderately hydrophobic peptide demonstrate greater selectivity than peptoids of increased hydrophobicity, due to the similar antimicrobial activity and diminished hemolytic activity.

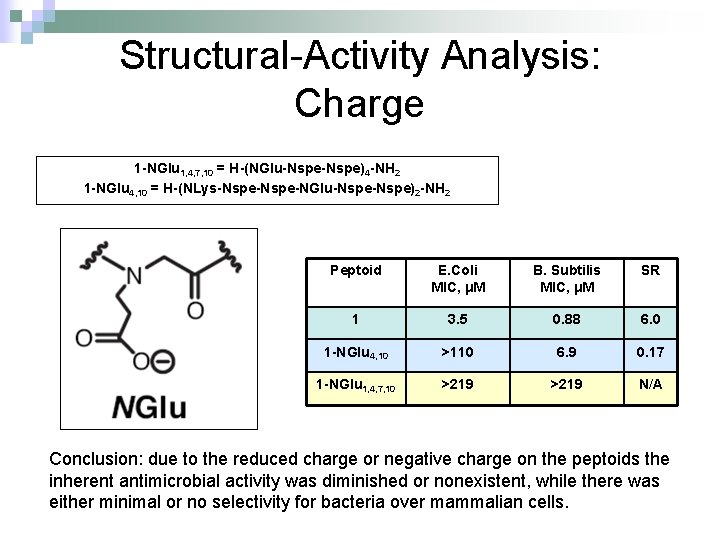
Structural-Activity Analysis: Charge 1 -NGlu 1, 4, 7, 10 = H-(NGlu-Nspe)4 -NH 2 1 -NGlu 4, 10 = H-(NLys-Nspe-NGlu-Nspe)2 -NH 2 Peptoid E. Coli MIC, μM B. Subtilis MIC, μM SR 1 3. 5 0. 88 6. 0 1 -NGlu 4, 10 >110 6. 9 0. 17 1 -NGlu 1, 4, 7, 10 >219 N/A Conclusion: due to the reduced charge or negative charge on the peptoids the inherent antimicrobial activity was diminished or nonexistent, while there was either minimal or no selectivity for bacteria over mammalian cells.

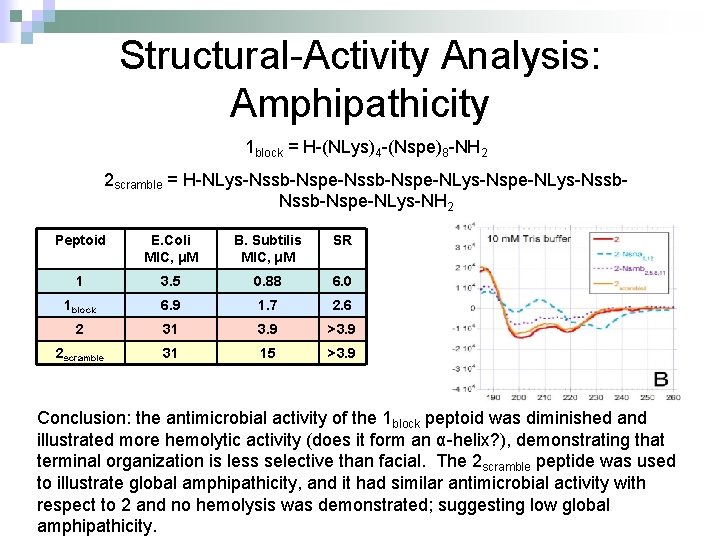
Structural-Activity Analysis: Amphipathicity 1 block = H-(NLys)4 -(Nspe)8 -NH 2 2 scramble = H-NLys-Nssb-Nspe-NLys-Nspe-NLys-Nssb-Nspe-NLys-NH 2 Peptoid E. Coli MIC, μM B. Subtilis MIC, μM SR 1 3. 5 0. 88 6. 0 1 block 6. 9 1. 7 2. 6 2 31 3. 9 >3. 9 2 scramble 31 15 >3. 9 Conclusion: the antimicrobial activity of the 1 block peptoid was diminished and illustrated more hemolytic activity (does it form an α-helix? ), demonstrating that terminal organization is less selective than facial. The 2 scramble peptide was used to illustrate global amphipathicity, and it had similar antimicrobial activity with respect to 2 and no hemolysis was demonstrated; suggesting low global amphipathicity.

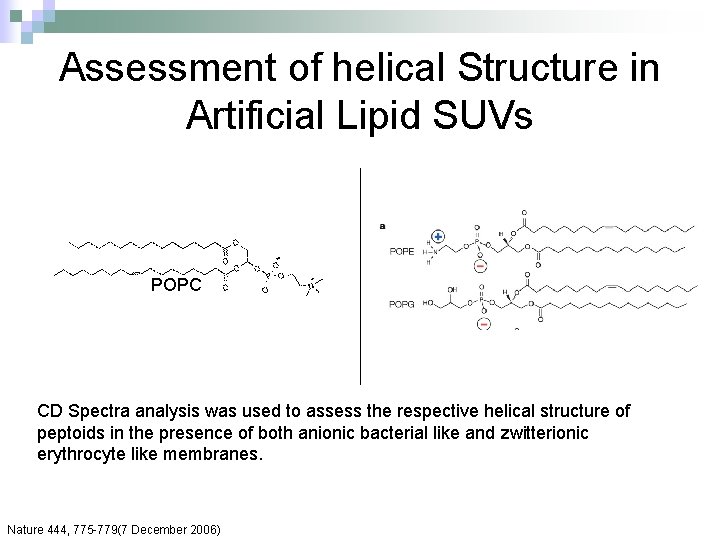
Assessment of helical Structure in Artificial Lipid SUVs POPC CD Spectra analysis was used to assess the respective helical structure of peptoids in the presence of both anionic bacterial like and zwitterionic erythrocyte like membranes. Nature 444, 775 -779(7 December 2006)

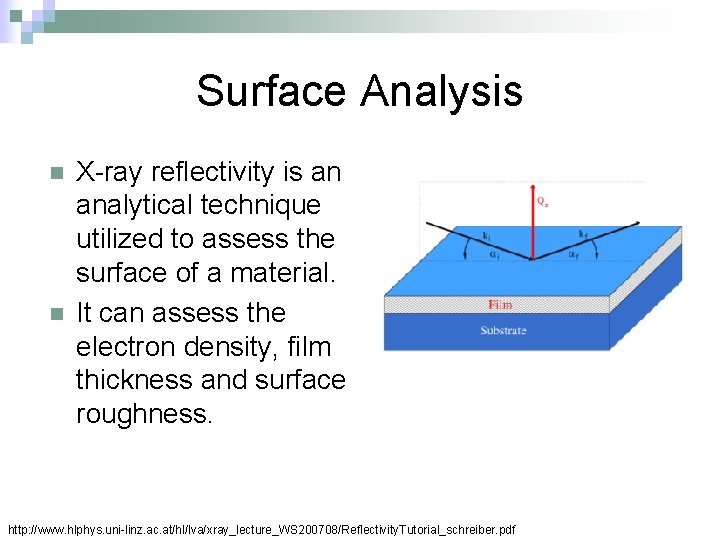
Surface Analysis n n X-ray reflectivity is an analytical technique utilized to assess the surface of a material. It can assess the electron density, film thickness and surface roughness. http: //www. hlphys. uni-linz. ac. at/hl/lva/xray_lecture_WS 200708/Reflectivity. Tutorial_schreiber. pdf

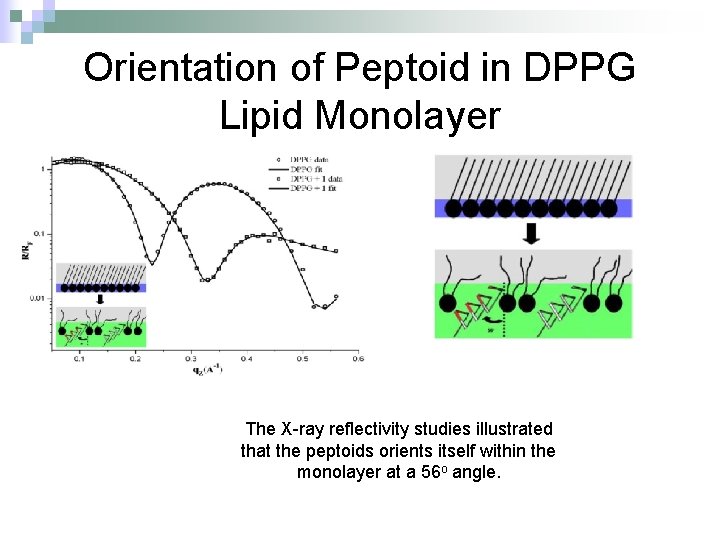
Orientation of Peptoid in DPPG Lipid Monolayer The X-ray reflectivity studies illustrated that the peptoids orients itself within the monolayer at a 56 o angle.

Conclusions 1. 2. 3. 4. Antimicrobial peptoids (amptoids) can function like antimicrobial peptides. They are selective for bacteria if they posses a cationic charge and are moderately hydrophobic. A defined helical structure is not required for antimicrobial activity, rather hemolytic activity. The peptoids interact with the membrane by inserting itself into the lipid layer – as the xray data suggests. However, the mechanism of action cannot be deduced at this time, but is similar to that of anitmicrobial peptides.

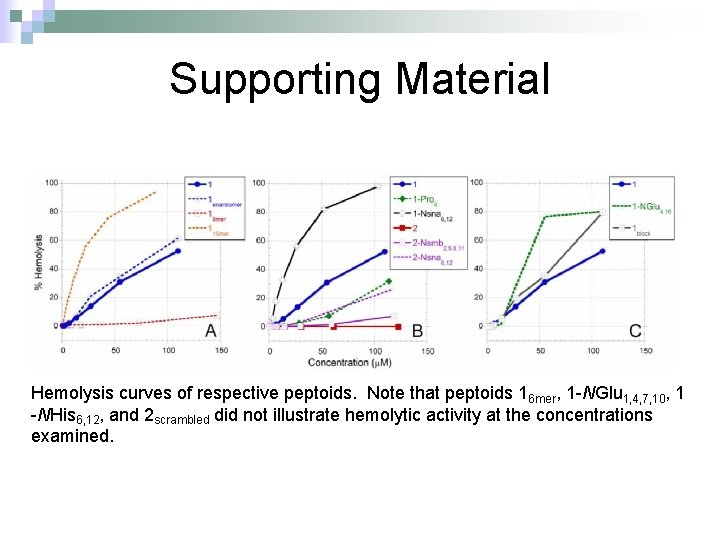
Supporting Material Hemolysis curves of respective peptoids. Note that peptoids 16 mer, 1 -NGlu 1, 4, 7, 10, 1 -NHis 6, 12, and 2 scrambled did not illustrate hemolytic activity at the concentrations examined.

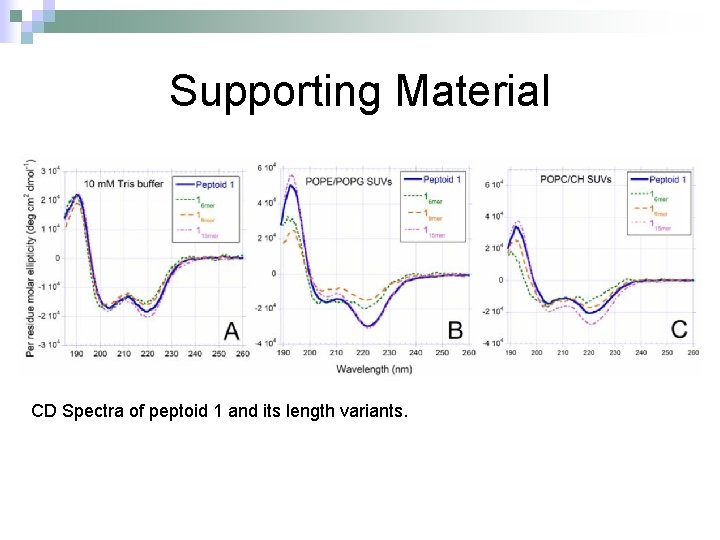
Supporting Material CD Spectra of peptoid 1 and its length variants.

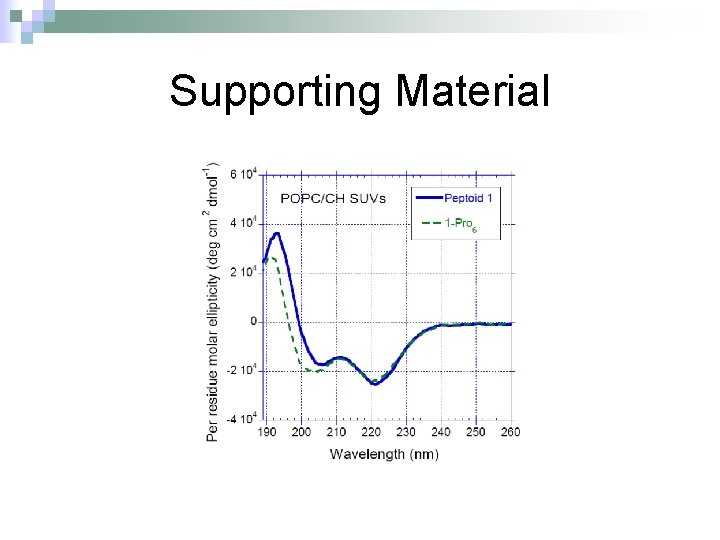
Supporting Material