Peptide Bond Dihedral Angles Ramachandran Plot Protein Data
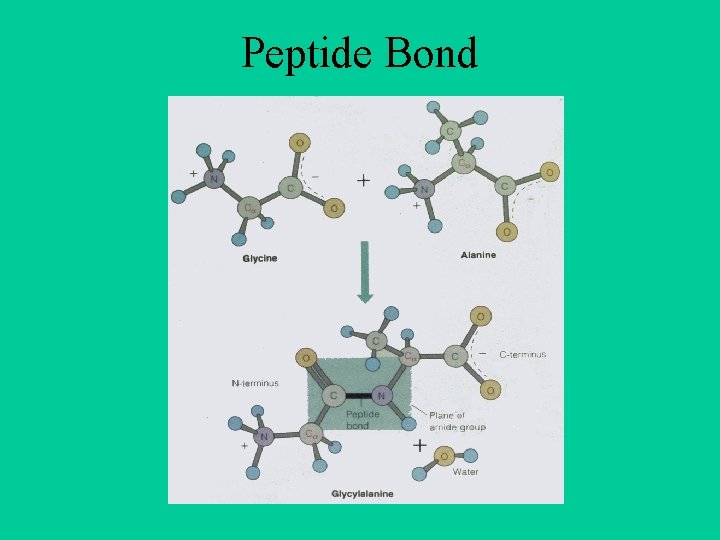
Peptide Bond
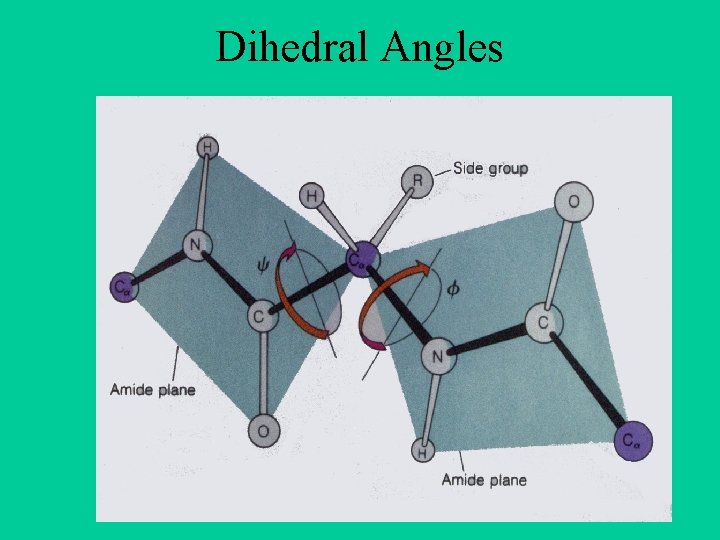
Dihedral Angles
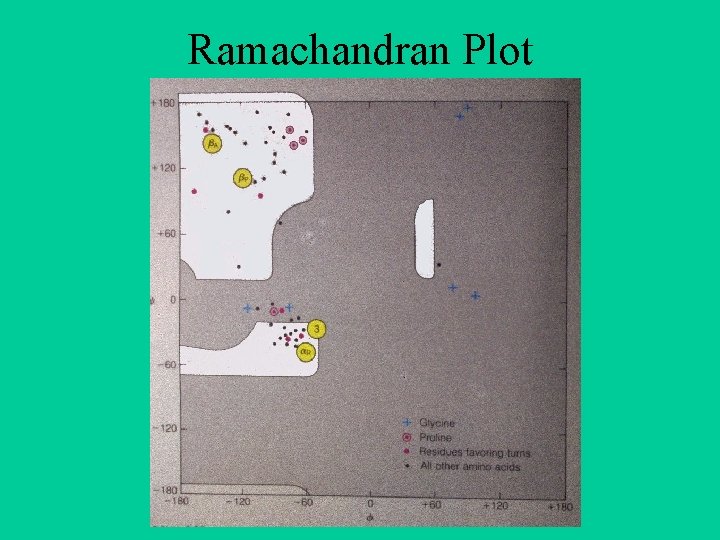
Ramachandran Plot
Protein Data. Bank (PDB) • Important in solving real problems in molecular biology • Protein Databank – PDB Established in 1972 at Brookhaven National Laboratory (BNL) – Sole international repository of macromolecular structure data – Moved to Research Collaboratory for Structural Bioinformatics http: //www. rcsb. org/

PDB: example HEADER LYASE(OXO-ACID) 01 -OCT-91 12 CA 2 COMPND CARBONIC ANHYDRASE /II (CARBONATE DEHYDRATASE) (/HCA II) 12 CA 3 COMPND 2 (E. C. 4. 2. 1. 1) MUTANT WITH VAL 121 REPLACED BY ALA (/V 121 A) 12 CA 4 SOURCE HUMAN (HOMO SAPIENS) RECOMBINANT PROTEIN 12 CA 5 AUTHOR S. K. NAIR, D. W. CHRISTIANSON 12 CA 6 REVDAT 1 15 -OCT-92 12 CA 0 12 CA 7 JRNL AUTH S. K. NAIR, T. L. CALDERONE, D. W. CHRISTIANSON, C. A. FIERKE 12 CA 8 JRNL TITL ALTERING THE MOUTH OF A HYDROPHOBIC POCKET. 12 CA 9 JRNL TITL 2 STRUCTURE AND KINETICS OF HUMAN CARBONIC ANHYDRASE 12 CA 10 JRNL TITL 3 /II$ MUTANTS AT RESIDUE VAL-121 12 CA 11 JRNL REF J. BIOL. CHEM. V. 266 17320 1991 12 CA 12 JRNL REFN ASTM JBCHA 3 US ISSN 0021 -9258 071 12 CA 13 REMARK 1 12 CA 14 REMARK 2 12 CA 15 REMARK 2 RESOLUTION. 2. 4 ANGSTROMS. 12 CA 16 REMARK 3 12 CA 17 REMARK 3 REFINEMENT. 12 CA 18 REMARK 3 PROGRAM PROLSQ 12 CA 19 REMARK 3 AUTHORS HENDRICKSON, KONNERT 12 CA 20 REMARK 3 R VALUE 0. 170 12 CA 21 REMARK 3 RMSD BOND DISTANCES 0. 011 ANGSTROMS 12 CA 22 REMARK 3 RMSD BOND ANGLES 1. 3 DEGREES 12 CA 23 REMARK 4 12 CA 24 REMARK 4 N-TERMINAL RESIDUES SER 2, HIS 3, HIS 4 AND C-TERMINAL 12 CA 25 REMARK 4 RESIDUE LYS 260 WERE NOT LOCATED IN THE DENSITY MAPS AND, 12 CA 26
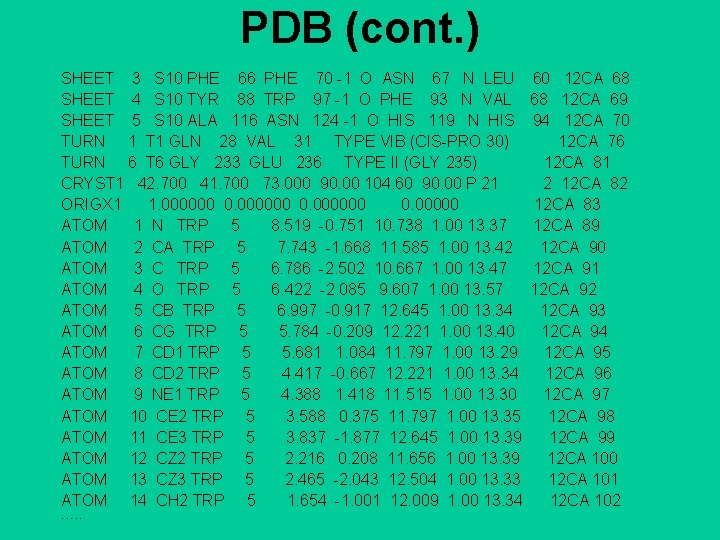
PDB (cont. ) SHEET 3 S 10 PHE 66 PHE 70 -1 O ASN 67 N LEU SHEET 4 S 10 TYR 88 TRP 97 -1 O PHE 93 N VAL SHEET 5 S 10 ALA 116 ASN 124 -1 O HIS 119 N HIS TURN 1 T 1 GLN 28 VAL 31 TYPE VIB (CIS-PRO 30) TURN 6 T 6 GLY 233 GLU 236 TYPE II (GLY 235) CRYST 1 42. 700 41. 700 73. 000 90. 00 104. 60 90. 00 P 21 ORIGX 1 1. 000000 0. 00000 ATOM 1 N TRP 5 8. 519 -0. 751 10. 738 1. 00 13. 37 ATOM 2 CA TRP 5 7. 743 -1. 668 11. 585 1. 00 13. 42 ATOM 3 C TRP 5 6. 786 -2. 502 10. 667 1. 00 13. 47 ATOM 4 O TRP 5 6. 422 -2. 085 9. 607 1. 00 13. 57 ATOM 5 CB TRP 5 6. 997 -0. 917 12. 645 1. 00 13. 34 ATOM 6 CG TRP 5 5. 784 -0. 209 12. 221 1. 00 13. 40 ATOM 7 CD 1 TRP 5 5. 681 1. 084 11. 797 1. 00 13. 29 ATOM 8 CD 2 TRP 5 4. 417 -0. 667 12. 221 1. 00 13. 34 ATOM 9 NE 1 TRP 5 4. 388 1. 418 11. 515 1. 00 13. 30 ATOM 10 CE 2 TRP 5 3. 588 0. 375 11. 797 1. 00 13. 35 ATOM 11 CE 3 TRP 5 3. 837 -1. 877 12. 645 1. 00 13. 39 ATOM 12 CZ 2 TRP 5 2. 216 0. 208 11. 656 1. 00 13. 39 ATOM 13 CZ 3 TRP 5 2. 465 -2. 043 12. 504 1. 00 13. 33 ATOM 14 CH 2 TRP 5 1. 654 -1. 001 12. 009 1. 00 13. 34 ……. 60 12 CA 68 68 12 CA 69 94 12 CA 70 12 CA 76 12 CA 81 2 12 CA 83 12 CA 89 12 CA 90 12 CA 91 12 CA 92 12 CA 93 12 CA 94 12 CA 95 12 CA 96 12 CA 97 12 CA 98 12 CA 99 12 CA 100 12 CA 101 12 CA 102
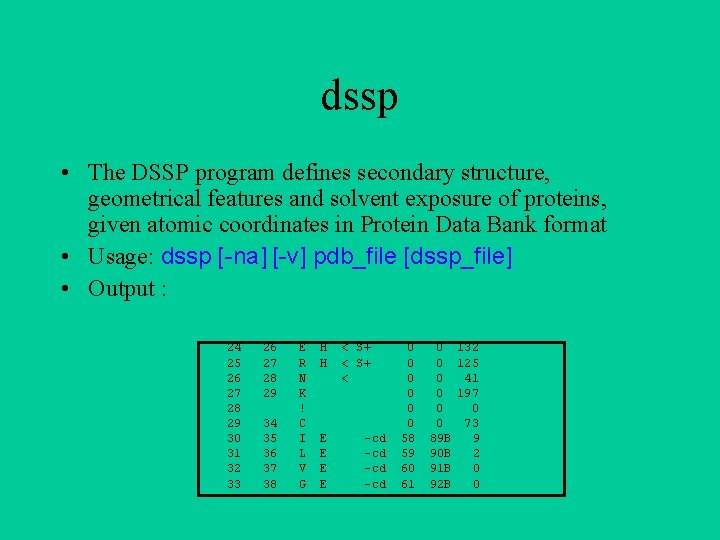
dssp • The DSSP program defines secondary structure, geometrical features and solvent exposure of proteins, given atomic coordinates in Protein Data Bank format • Usage: dssp [-na] [-v] pdb_file [dssp_file] • Output : 24 25 26 27 28 29 30 31 32 33 26 27 28 29 34 35 36 37 38 E R N K ! C I L V G H H E E < S+ < -cd -cd 0 0 0 58 59 60 61 0 132 0 125 0 41 0 197 0 0 0 73 89 B 9 90 B 2 91 B 0 92 B 0

Techniques of Structure Prediction • Computer simulation based on energy calculation – Based on physio-chemical principles – Thermodynamic equilibrium with a minimum free energy – Global minimum free energy of protein surface • Knowledge Based approaches – Homology Based Approach – Threading Protein Sequence – Hierarchical Methods

Energy Minimization Techniques Energy Minimization based methods in their pure form, make no priori assumptions and attempt to locate global minma. • Static Minimization Methods – Classical many potential-potential can be construted – Assume that atoms in protein is in static form – Problems(large number of variables & minima and validity of potentials) • Dynamical Minimization Methods – Motions of atoms also considered – Monte Carlo simulation (stochastics in nature, time is not consider) – Molecular Dynamics (time, quantum mechanical, classical equ. ) • Limitations – large number of degree of freedom, CPU power not adequate – Interaction potential is not good enough to model
Knowledge based Techniques • Homology Modelling – Need homologues of known protein structure – Backbone modelling – Side chain modelling – Fail in absence of homology • Threading Based Methods – New way of fold recognition – Sequence is tried to fit in known structures – Motif recognition – Loop & Side chain modelling – Fail in absence of known example

Hierarchical Methods Intermidiate structures are predicted, instead of predicting tertiary structure of protein from amino acids sequence • Prediction of backbone structure – Secondary structure (helix, sheet, coil) – Beta Turn Prediction – Super-secondary structure • Tertiary structure prediction • Limitation Accuracy is only 75 -80 % Only three state prediction
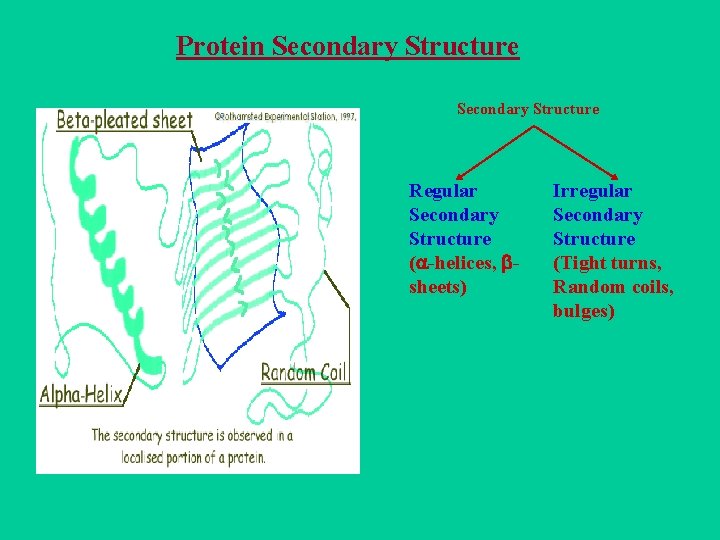
Protein Secondary Structure Regular Secondary Structure ( -helices, sheets) Irregular Secondary Structure (Tight turns, Random coils, bulges)
Secondary structure prediction No information about tight turns ?
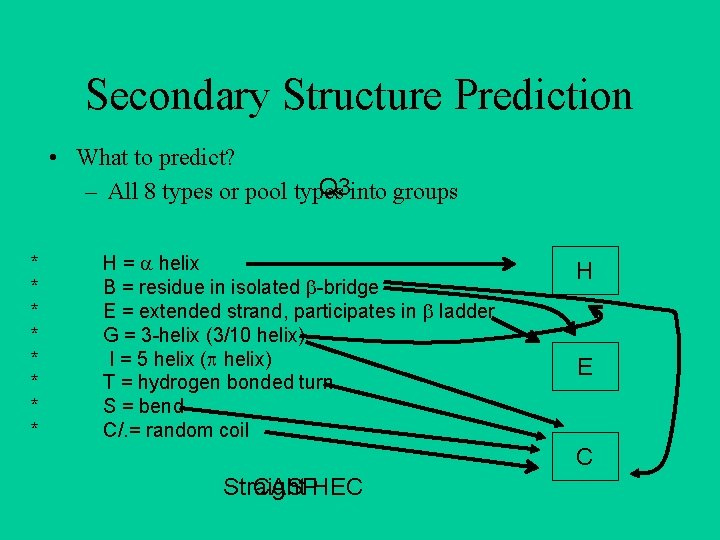
Secondary Structure Prediction • What to predict? Q 3 into groups – All 8 types or pool types * * * * H = helix B = residue in isolated -bridge E = extended strand, participates in ladder G = 3 -helix (3/10 helix) I = 5 helix ( helix) T = hydrogen bonded turn S = bend C/. = random coil H E C Straight CASPHEC
Type of Secondary Structure Prediction • Information based classification – – Property based methods (Manual / Subjective) Residue based methods Segment or peptide based approaches Application of Multiple Sequence Alignment • Technical classification – Statistical Methods • Chou & fashman (1974) • GOR – Artificial Itellegence Based Methods • • Neural Network Based Methods (1988) Nearest Neighbour Methods (1992) Hidden Markove model (1993) Support Vector Machine based methods
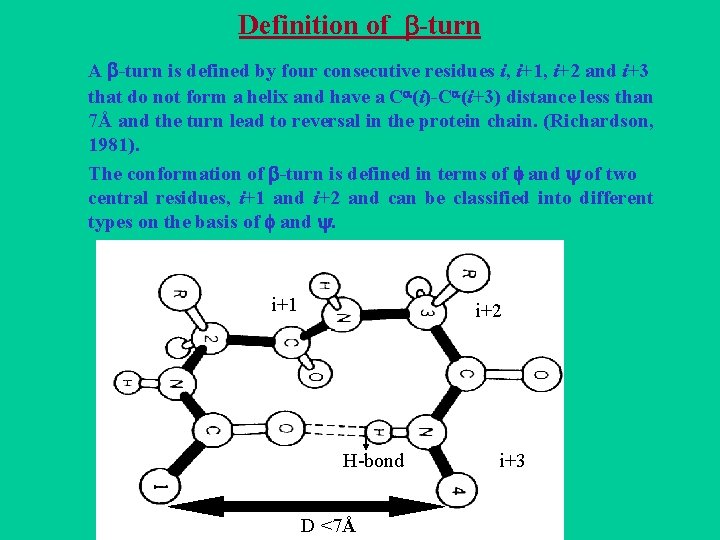
Definition of -turn A -turn is defined by four consecutive residues i, i+1, i+2 and i+3 that do not form a helix and have a C (i)-C (i+3) distance less than 7Å and the turn lead to reversal in the protein chain. (Richardson, 1981). The conformation of -turn is defined in terms of and of two central residues, i+1 and i+2 and can be classified into different types on the basis of and . i+1 i i+2 H-bond D <7Å i+3
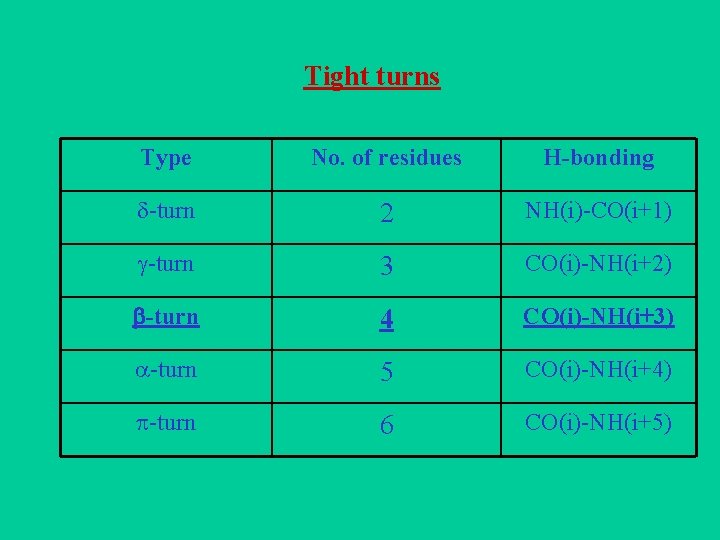
Tight turns Type No. of residues H-bonding -turn 2 NH(i)-CO(i+1) -turn 3 CO(i)-NH(i+2) -turn 4 CO(i)-NH(i+3) -turn 5 CO(i)-NH(i+4) -turn 6 CO(i)-NH(i+5)

Prediction of tight turns • • • Prediction of -turns Prediction of -turn types Prediction of -turns Use the tight turns information, mainly -turns in tertiary structure prediction of bioactive peptides

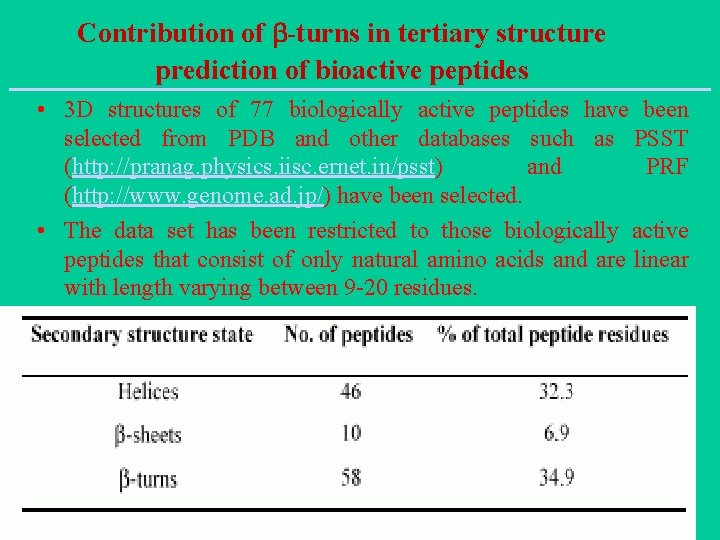
Contribution of -turns in tertiary structure prediction of bioactive peptides • 3 D structures of 77 biologically active peptides have been selected from PDB and other databases such as PSST (http: //pranag. physics. iisc. ernet. in/psst) and PRF (http: //www. genome. ad. jp/) have been selected. • The data set has been restricted to those biologically active peptides that consist of only natural amino acids and are linear with length varying between 9 -20 residues.
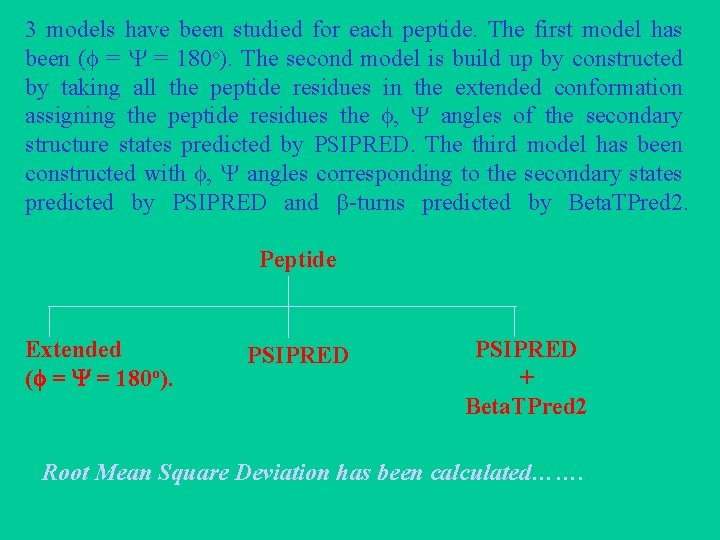
3 models have been studied for each peptide. The first model has been ( = = 180 o). The second model is build up by constructed by taking all the peptide residues in the extended conformation assigning the peptide residues the , angles of the secondary structure states predicted by PSIPRED. The third model has been constructed with , angles corresponding to the secondary states predicted by PSIPRED and -turns predicted by Beta. TPred 2. Peptide Extended ( = = 180 o). PSIPRED + Beta. TPred 2 Root Mean Square Deviation has been calculated…….
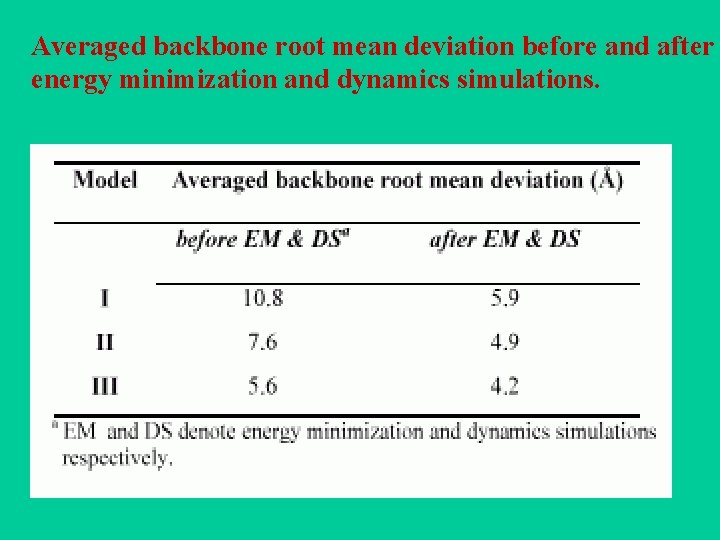
Averaged backbone root mean deviation before and after energy minimization and dynamics simulations.
Protein Structure Prediction • Regular Secondary Structure Prediction ( -helix -sheet) – APSSP 2: Highly accurate method for secondary structure prediction – Participate in all competitions like EVA, CAFASP and CASP (In top 5 methods) – Combines memory based reasoning ( MBR) and ANN methods • Irregular secondary structure prediction methods (Tight turns) – Betatpred: Consensus method for -turns prediction • Statistical methods combined • Kaur and Raghava (2001) Bioinformatics – Bteval : Benchmarking of -turns prediction • Kaur and Raghava (2002) J. Bioinformatics and Computational Biology, 1: 495: 504 – Beta. Tpred 2: Highly accurate method for predicting -turns (ANN, SS, MA) • Multiple alignment and secondary structure information • Kaur and Raghava (2003) Protein Sci 12: 627 -34 – Beta. Turns: Prediction of -turn types in proteins • Evolutionary information • Kaur and Raghava (2004) Bioinformatics 20: 2751 -8. – Alpha. Pred: Prediction of -turns in proteins • Kaur and Raghava (2004) Proteins: Structure, Function, and Genetics 55: 83 -90 – Gamma. Pred: Prediction of -turns in proteins • Kaur and Raghava (2004) Protein Science; 12: 923 -929.
Protein Structure Prediction • Bhair. Pred: Prediction of Supersecondary structure prediction – – • TBBpred: Prediction of outer membrane proteins – – • • Prediction of trans membrane beta barrel proteins Prediction of beta barrel regions Application of ANN and SVM + Evolutionary information Natt et al. (2004) Proteins: 56: 11 -8 ARNHpred: Analysis and prediction side chain, backbone interactions – Prediction of aromatic NH interactions – Kaur and Raghava (2004) FEBS Letters 564: 47 -57. SARpred: Prediction of surface accessibility (real accessibility) – – – • Prediction of Beta Hairpins Utilize ANN and SVM pattern recognition techniques Secondary structure and surface accessibility used as input Manish et al. (2005) Nucleic Acids Research (In press) Multiple alignment (PSIBLAST) and Secondary structure information ANN: Two layered network (sequence-structure) Garg et al. , (2005) Proteins (In Press) Pep. Str: Prediction of tertiary structure of Bioactive peptides Performance of SARpred, Pepstr and Bhair. Pred were checked on CASP 6 proteins

Thankyou
- Slides: 25