Peak Seq Rozowsky 2009 Peak Seq Updates The
![Peak. Seq: [Rozowsky, 2009] Peak. Seq: [Rozowsky, 2009]](https://slidetodoc.com/presentation_image_h/28fd83ce5e07ef024e98b55537301da9/image-1.jpg)


- Slides: 6
![Peak Seq Rozowsky 2009 Peak. Seq: [Rozowsky, 2009]](https://slidetodoc.com/presentation_image_h/28fd83ce5e07ef024e98b55537301da9/image-1.jpg)
Peak. Seq: [Rozowsky, 2009]

Peak. Seq: Updates • The code is re-organized and re-written • Supports multiple input read formats – SAM, ELAND, Default BOWTIE format, tag. Align – Supports BAM via piping SAM output from samtools • Easier to setup than previous version • Works faster: – Observed that peak calling on the Pol 2 dataset on the website finishes in half the time • There is also new code for setting up Peak. Seq for peak calling on a large dataset • Available at: http: //archive. gersteinlab. org/proj/Peak. Seq/Scoring_Ch. IPSeq/Code/C/

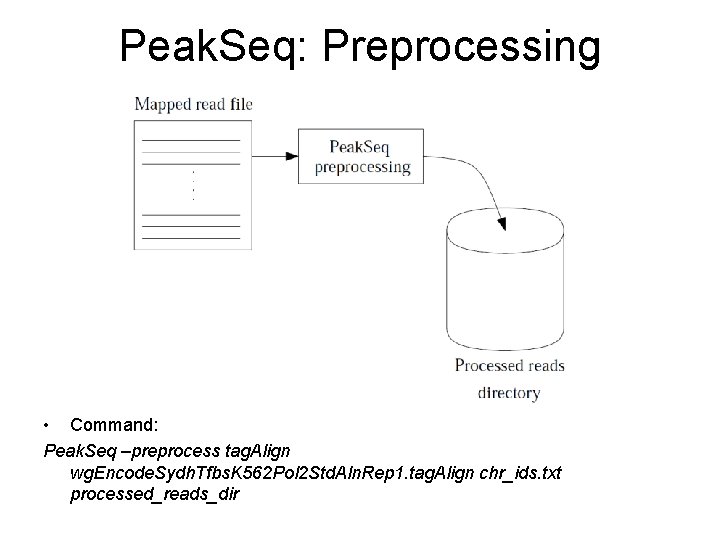
Peak. Seq: Preprocessing • Command: Peak. Seq –preprocess tag. Align wg. Encode. Sydh. Tfbs. K 562 Pol 2 Std. Aln. Rep 1. tag. Align chr_ids. txt processed_reads_dir

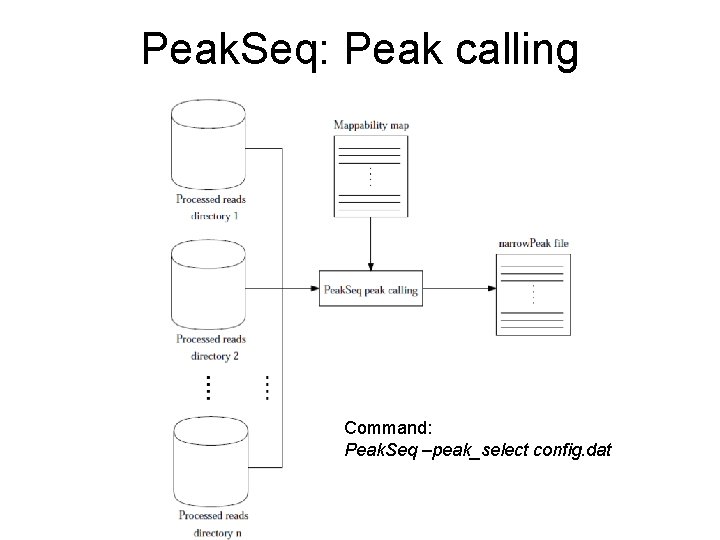
Peak. Seq: Peak calling Command: Peak. Seq –peak_select config. dat

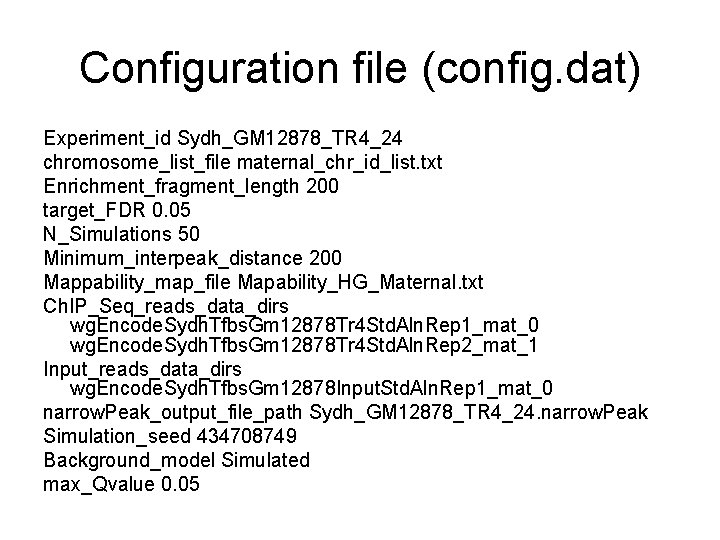
Configuration file (config. dat) Experiment_id Sydh_GM 12878_TR 4_24 chromosome_list_file maternal_chr_id_list. txt Enrichment_fragment_length 200 target_FDR 0. 05 N_Simulations 50 Minimum_interpeak_distance 200 Mappability_map_file Mapability_HG_Maternal. txt Ch. IP_Seq_reads_data_dirs wg. Encode. Sydh. Tfbs. Gm 12878 Tr 4 Std. Aln. Rep 1_mat_0 wg. Encode. Sydh. Tfbs. Gm 12878 Tr 4 Std. Aln. Rep 2_mat_1 Input_reads_data_dirs wg. Encode. Sydh. Tfbs. Gm 12878 Input. Std. Aln. Rep 1_mat_0 narrow. Peak_output_file_path Sydh_GM 12878_TR 4_24. narrow. Peak Simulation_seed 434708749 Background_model Simulated max_Qvalue 0. 05

Peak. Seq: Background models • There is an experimental Poisson background – Specified by setting “Background” entry to “Poisson” – Sets the threshold for each window to the average expected read depth for that window – Generates a relaxed threshold and lacks the target FDR requirement