PCR Polymerase Chain Reaction PCR is an in




- Slides: 23

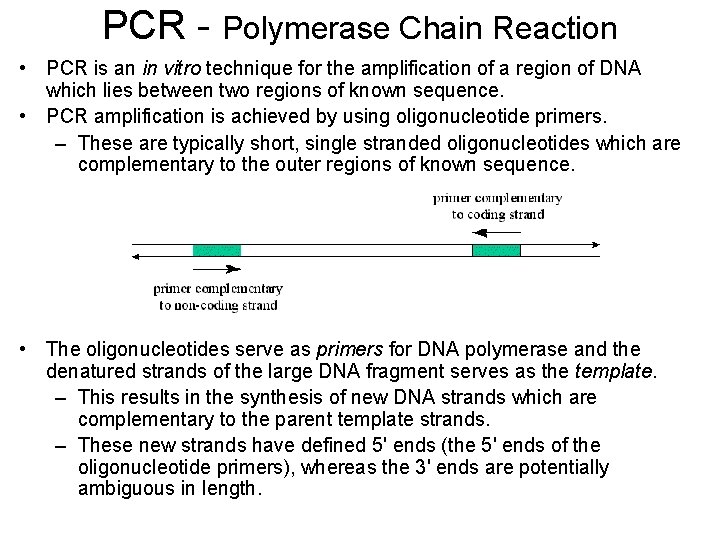
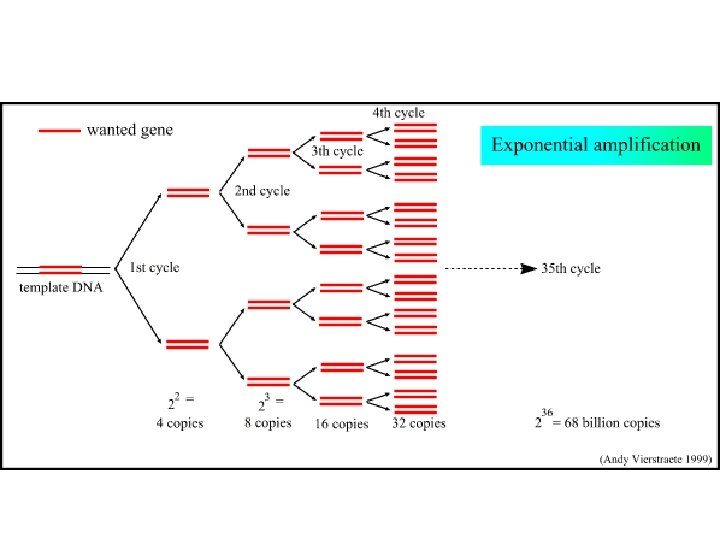
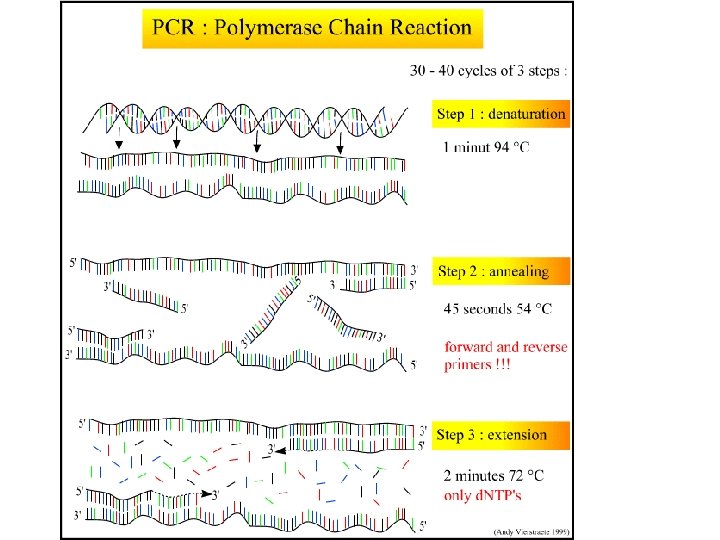
PCR - Polymerase Chain Reaction • PCR is an in vitro technique for the amplification of a region of DNA which lies between two regions of known sequence. • PCR amplification is achieved by using oligonucleotide primers. – These are typically short, single stranded oligonucleotides which are complementary to the outer regions of known sequence. • The oligonucleotides serve as primers for DNA polymerase and the denatured strands of the large DNA fragment serves as the template. – This results in the synthesis of new DNA strands which are complementary to the parent template strands. – These new strands have defined 5' ends (the 5' ends of the oligonucleotide primers), whereas the 3' ends are potentially ambiguous in length.



• http: //ocw. mit. edu/NR/rdonlyres/Civil-and-Environmental-Engineering/1 -89 Fall-2004/321 BF 8 FF-75 BE-4377 -8 D 74 -8 EEE 753 A 328 C/0/11_02_04. pdf

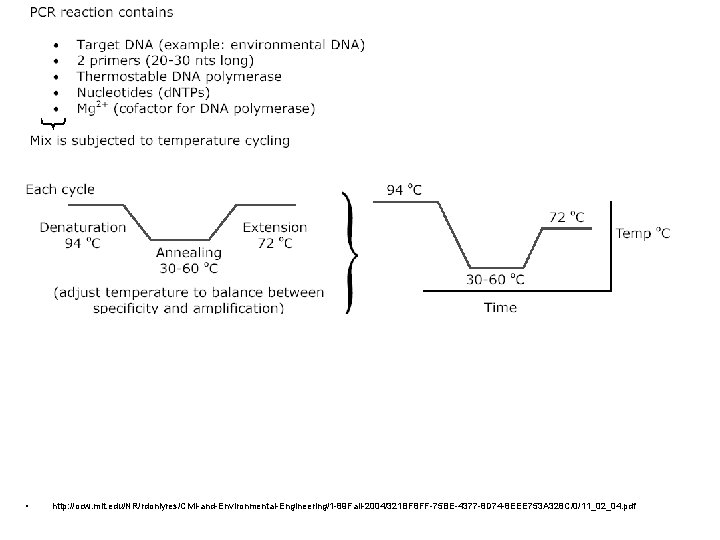
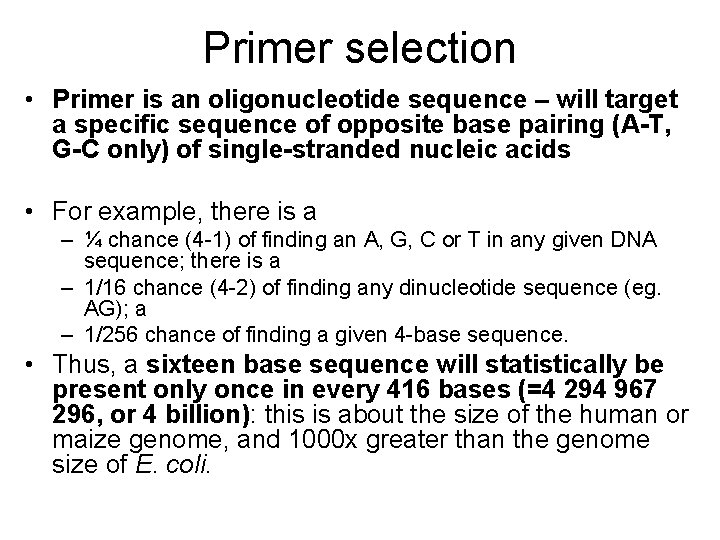
Primer selection • Primer is an oligonucleotide sequence – will target a specific sequence of opposite base pairing (A-T, G-C only) of single-stranded nucleic acids • For example, there is a – ¼ chance (4 -1) of finding an A, G, C or T in any given DNA sequence; there is a – 1/16 chance (4 -2) of finding any dinucleotide sequence (eg. AG); a – 1/256 chance of finding a given 4 -base sequence. • Thus, a sixteen base sequence will statistically be present only once in every 416 bases (=4 294 967 296, or 4 billion): this is about the size of the human or maize genome, and 1000 x greater than the genome size of E. coli.

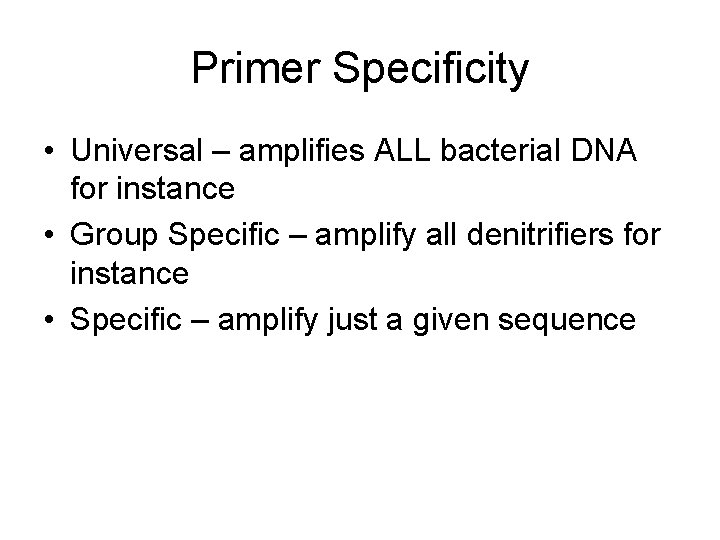
Primer Specificity • Universal – amplifies ALL bacterial DNA for instance • Group Specific – amplify all denitrifiers for instance • Specific – amplify just a given sequence

Forward and reverse primers • If you know the sequence targeted for amplification, you know the size which the primers should be anealing across • If you don’t know the sequence… What do you get?

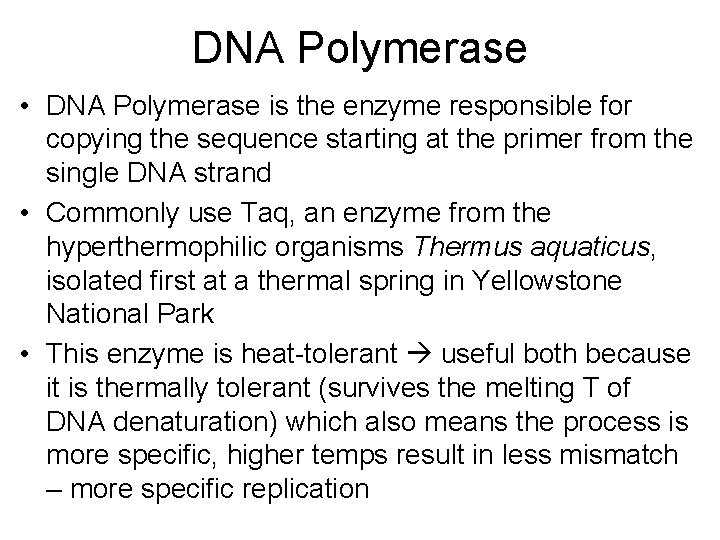
DNA Polymerase • DNA Polymerase is the enzyme responsible for copying the sequence starting at the primer from the single DNA strand • Commonly use Taq, an enzyme from the hyperthermophilic organisms Thermus aquaticus, isolated first at a thermal spring in Yellowstone National Park • This enzyme is heat-tolerant useful both because it is thermally tolerant (survives the melting T of DNA denaturation) which also means the process is more specific, higher temps result in less mismatch – more specific replication

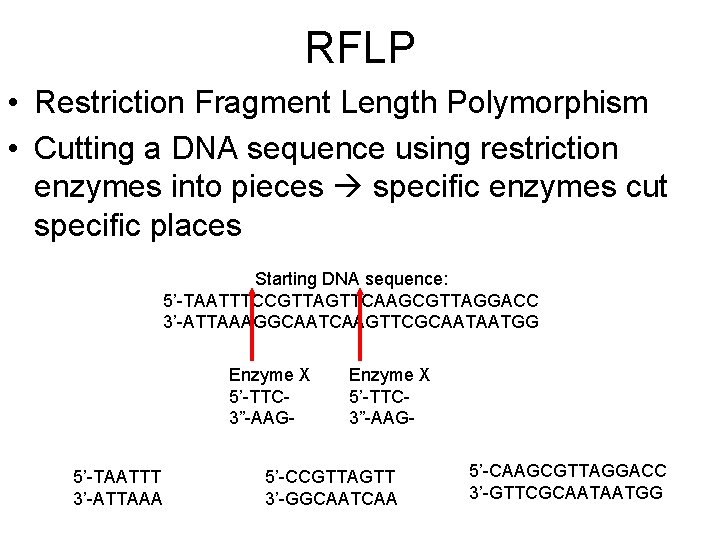
RFLP • Restriction Fragment Length Polymorphism • Cutting a DNA sequence using restriction enzymes into pieces specific enzymes cut specific places Starting DNA sequence: 5’-TAATTTCCGTTAGTTCAAGCGTTAGGACC 3’-ATTAAAGGCAATCAAGTTCGCAATAATGG Enzyme X 5’-TTC 3”-AAG 5’-TAATTT 3’-ATTAAA Enzyme X 5’-TTC 3”-AAG- 5’-CCGTTAGTT 3’-GGCAATCAA 5’-CAAGCGTTAGGACC 3’-GTTCGCAATAATGG

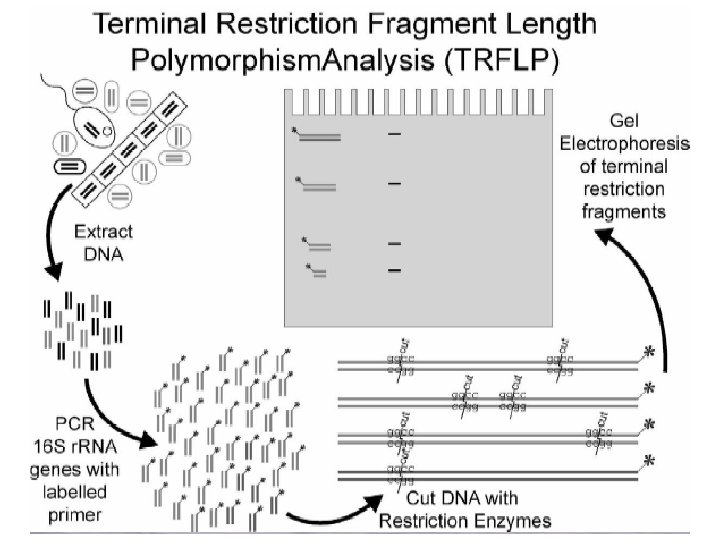
RFLP • DNA can be processed by RFLP either directly (if you can get enough DNA from an environment) or from PCR product • T-RFLP (terminal-RFLP) is in most respects identical except for a marker on the end of the enzyme • Works as fingerprinting technique because different organisms with different DNA sequences will have different lengths of DNA between identical units targeted by the restriction enzymes – specificity can again be manipulated with PCR primers Liu et al. (1997) Appl Environ Microbiol 63: 4516 -4522

Electrophoresis • Fragmentation products of differing length are separated – often on an agarose gel bed by electrophoresis, or using a capilarry electrophoretic separation


DGGE • Denaturing gradient gel electrophoresis – The hydrogen bonds formed between complimentary base pairs, GC rich regions ‘melt’ (melting=strand separation or denaturation) at higher temperatures than regions that are AT rich. • When DNA separated by electrophoresis through a gradient of increasing chemical denaturant (usually formamide and urea), the mobility of the molecule is retarded at the concentration at which the DNA strands of low melt domain dissociate. – The branched structure of the single stranded moiety of the molecule becomes entangled in the gel matrix and no further movement occurs. – Complete strand separation is prevented by the presence of a high melting domain, which is usually artificially created at one end of the molecule by incorporation of a GC clamp. This is accomplished during PCR amplification using a PCR primer with a 5' tail consisting of a sequence of 40 GC. Run DGGE animation here – from http: //www. charite. de/bioinf/tgge/

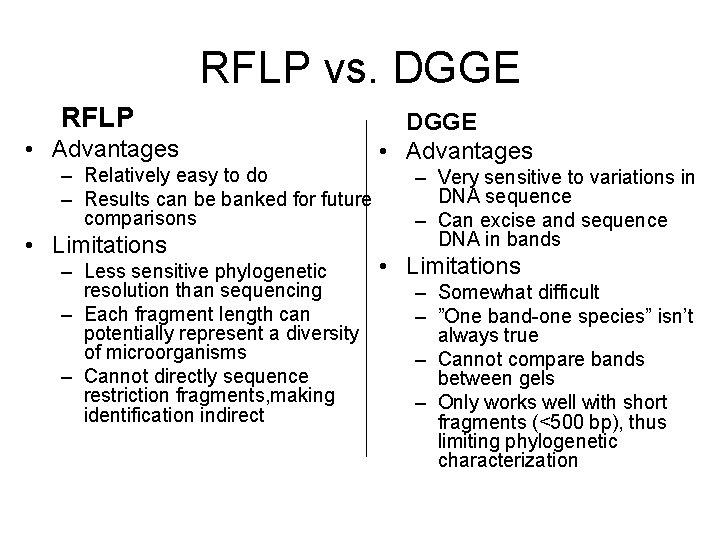
RFLP vs. DGGE RFLP • Advantages – Relatively easy to do – Results can be banked for future comparisons • Limitations DGGE • Advantages – Very sensitive to variations in DNA sequence – Can excise and sequence DNA in bands • Limitations – Less sensitive phylogenetic resolution than sequencing – Somewhat difficult – Each fragment length can – ”One band-one species” isn’t potentially represent a diversity always true of microorganisms – Cannot compare bands – Cannot directly sequence between gels restriction fragments, making – Only works well with short identification indirect fragments (<500 bp), thus limiting phylogenetic characterization

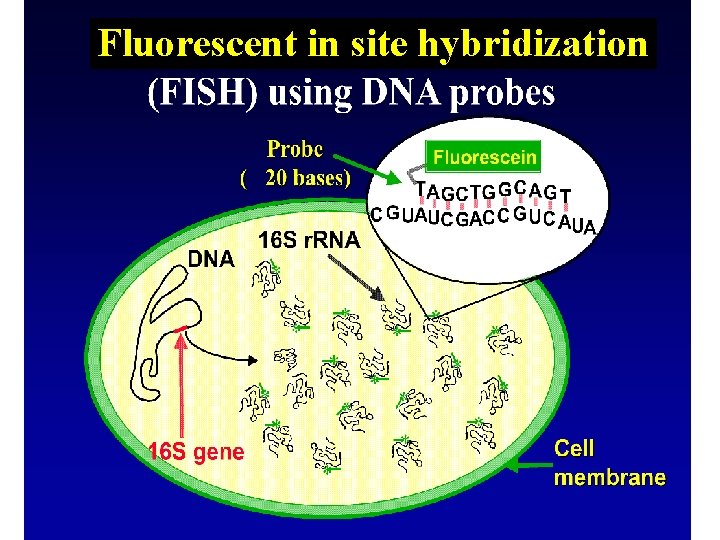
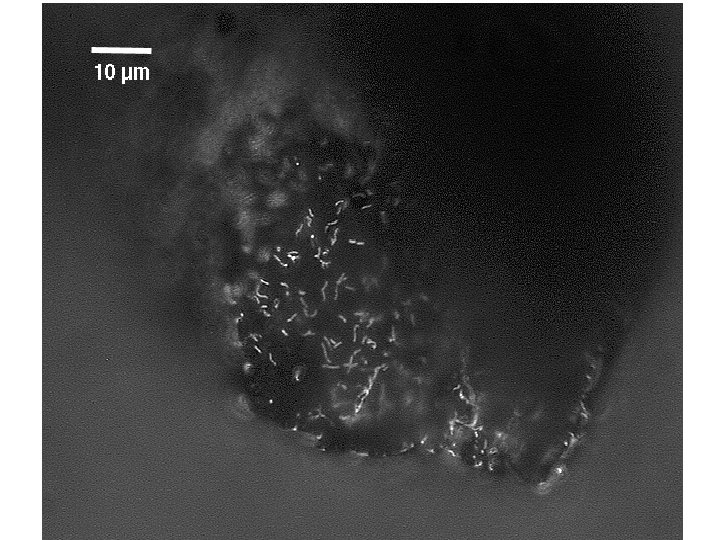
FISH • Fluorescent in-situ hybridization – Design a probe consisting of an oligonucleotide sequence and a tag – Degree of specificity is variable! – Hybridize that oligonucleotide sequence to the r. RNA of an organism – this is temperature and salt content sensitive – Image using epiflourescence, laser excitation confocal microscopy • Technique DIRECTLY images active organisms in a sample

Fluorescent in site hybridization

B Drift Slime Streamer 10 µm DAPI FER 656


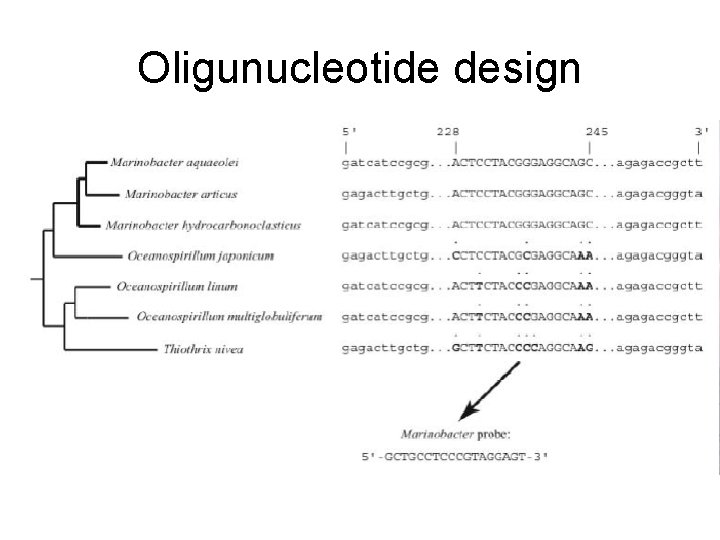
Oligunucleotide design

FISH variations • FISH-CARD – instead of a fluorescent probe on oligo sequence, but another molecule that can then bond to many fluorescent probes – better signal-to-noise ratio • FISH-RING – design of oligo sequence to specific genes – image all organisms with DSR gene or nif. H for example

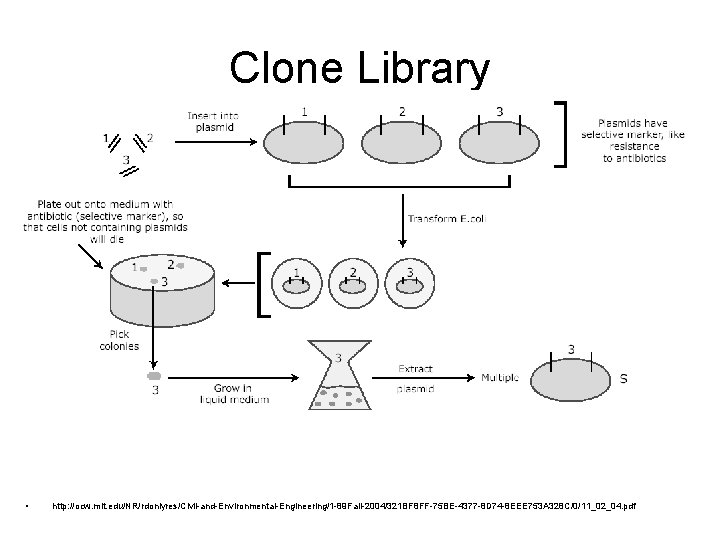
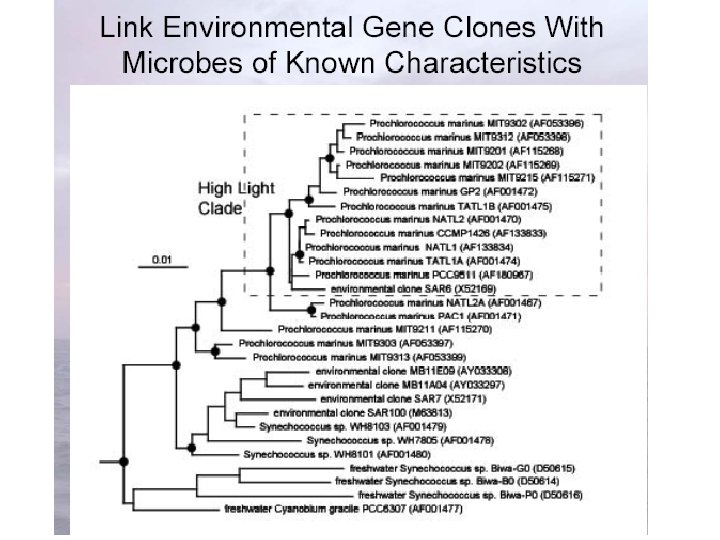
Clone Library • http: //ocw. mit. edu/NR/rdonlyres/Civil-and-Environmental-Engineering/1 -89 Fall-2004/321 BF 8 FF-75 BE-4377 -8 D 74 -8 EEE 753 A 328 C/0/11_02_04. pdf

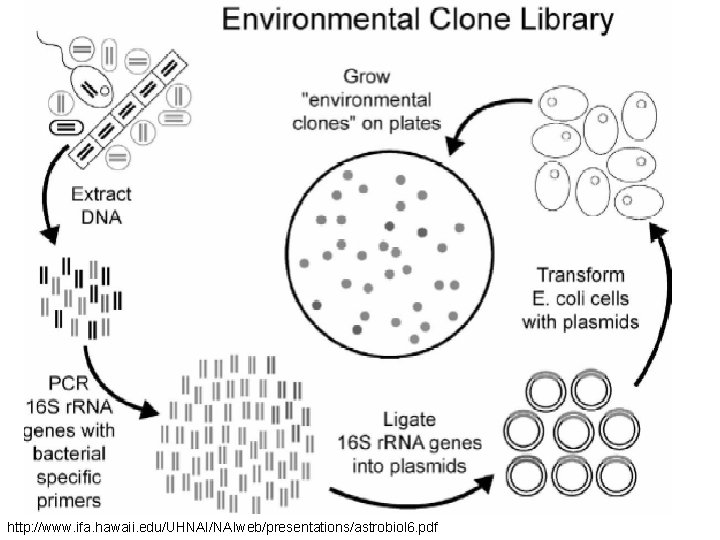
http: //www. ifa. hawaii. edu/UHNAI/NAIweb/presentations/astrobiol 6. pdf
