Parallelized Multiple Sequence Alignment on the Public Cloud
Parallelized Multiple Sequence Alignment on the Public Cloud Presented by: Dr. G. Sudha Sadasivam Professor, Dept of CSE, PSG College of Technology, Coimbatore Co-authors Mr B. Vijayan, Mr S. Arul Prakash, Mr K. V. Hari Babu Students, BE(CSE), Dept of CSE, PSG College of Technology, Coimbatore

Agenda § § § § § Sequence alignment Introduction to Clouds Approaches for MSA Problem statement System Architecture Illustration of working of the system Analysis Experimental results Conclusion
What is Sequence Alignment? The procedure of comparing two or more sequences by searching for a series of individual characters or character patterns that are in the same order in the sequences. § Uses § For sequence similarity § Phylogenetic tree analysis § Factors – accuracy and speed
Cloud computing Provides scalable, on-demand, RT computing services Suitability of cloud for Sequence Alignment § On-demand scalability of cloud makes it suitable for dynamic nature of MSA § Low cost in maintenance of infrastructure for applications § Data and compute parallelism in clouds through map-reduce paradigm facilitates energy efficient and fast MSA.
Types of Sequence Alignment Pair-wise Alignment of two sequences Global –using Needleman Wunsch algorithm. LGPSSKQTGKGS_SRAWDN | | | | LN_ATKSAGKGAIMRL GDA Local – using Smith Waterman algorithm. _____TGKG_____ | | | _____AGKG_____
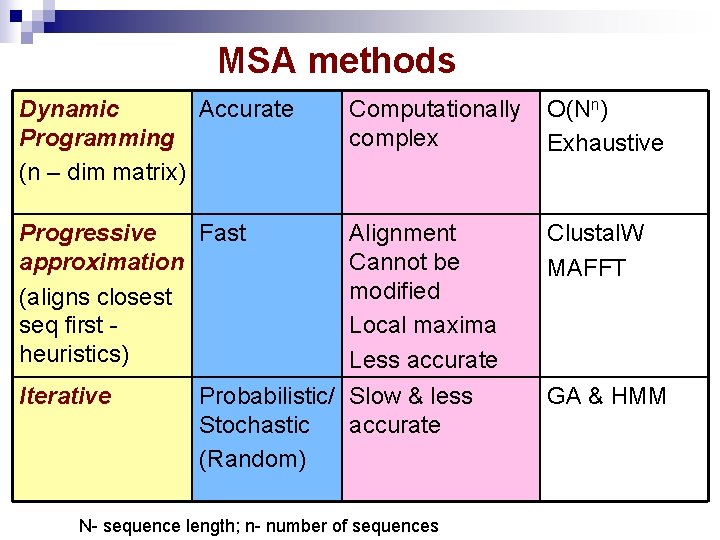
MSA methods Dynamic Accurate Programming (n – dim matrix) Computationally complex O(Nn) Exhaustive Progressive Fast approximation (aligns closest seq first heuristics) Alignment Cannot be modified Local maxima Less accurate Clustal. W MAFFT Iterative Probabilistic/ Slow & less Stochastic accurate (Random) N- sequence length; n- number of sequences GA & HMM
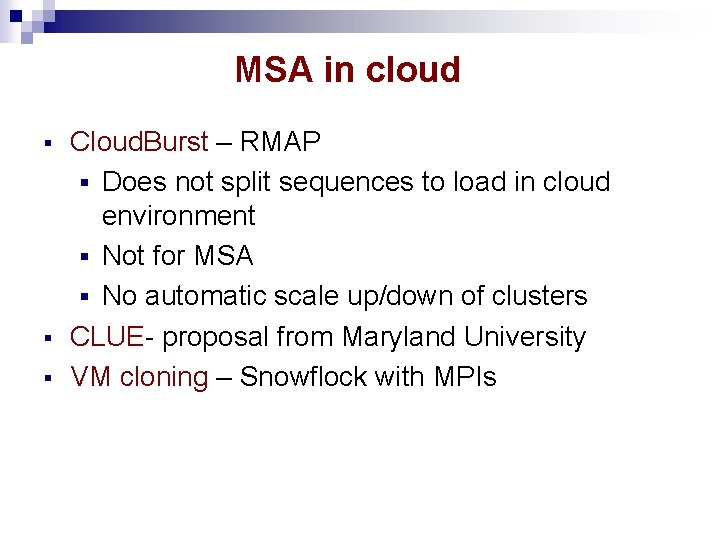
MSA in cloud § § § Cloud. Burst – RMAP § Does not split sequences to load in cloud environment § Not for MSA § No automatic scale up/down of clusters CLUE- proposal from Maryland University VM cloning – Snowflock with MPIs
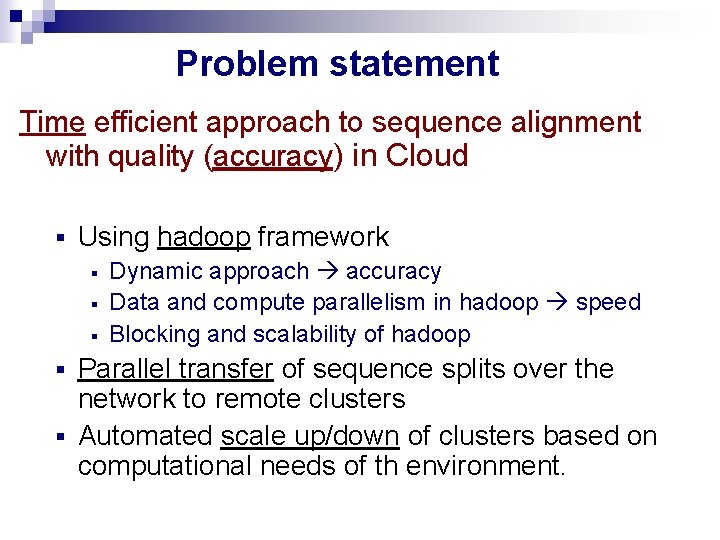
Problem statement Time efficient approach to sequence alignment with quality (accuracy) in Cloud § Using hadoop framework § § § Dynamic approach accuracy Data and compute parallelism in hadoop speed Blocking and scalability of hadoop Parallel transfer of sequence splits over the network to remote clusters § Automated scale up/down of clusters based on computational needs of th environment. §
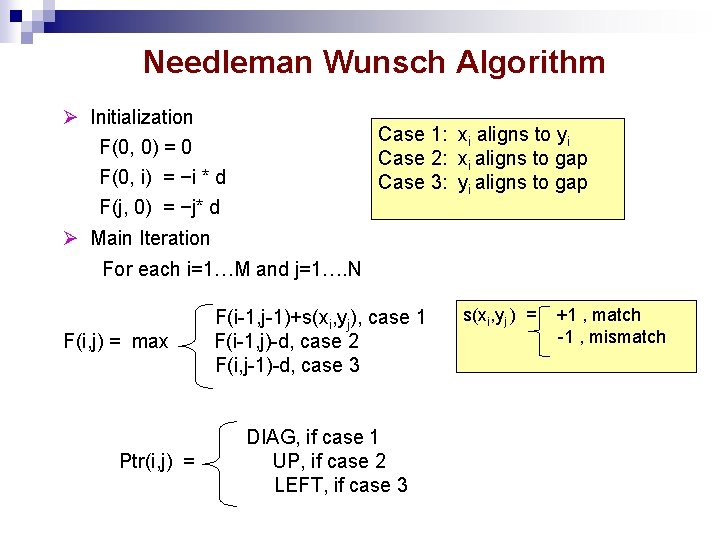
Needleman Wunsch Algorithm Initialization F(0, 0) = 0 F(0, i) = −i * d F(j, 0) = −j* d Case 1: xi aligns to yi Case 2: xi aligns to gap Case 3: yi aligns to gap Main Iteration For each i=1…M and j=1…. N F(i, j) = max Ptr(i, j) = F(i-1, j-1)+s(xi, yj), case 1 F(i-1, j)-d, case 2 F(i, j-1)-d, case 3 DIAG, if case 1 UP, if case 2 LEFT, if case 3 s(xi, yj ) = +1 , match -1 , mismatch
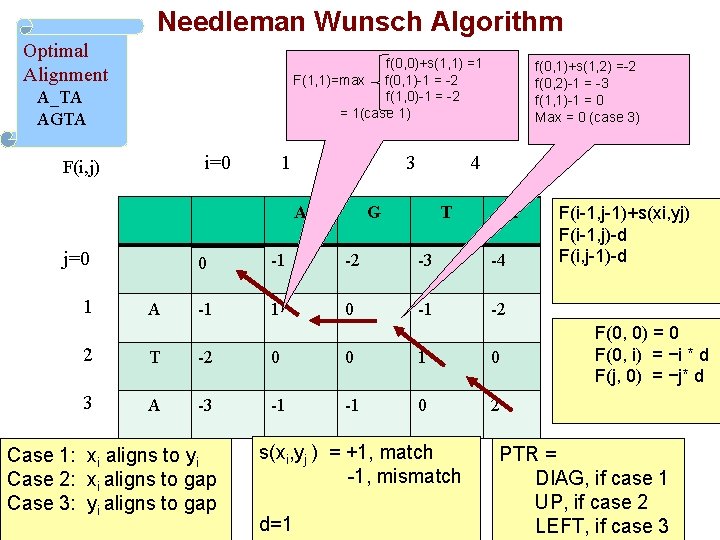
Needleman Wunsch Algorithm Optimal Alignment f(0, 0)+s(1, 1) =1 F(1, 1)=max f(0, 1)-1 = -2 f(1, 0)-1 = -2 = 1(case 1) A_TA AGTA i=0 F(i, j) 1 2 A j=0 1 A 3 f(0, 1)+s(1, 2) =-2 f(0, 2)-1 = -3 f(1, 1)-1 = 0 Max = 0 (case 3) 4 G T A 0 -1 -2 -3 -4 -1 1 0 -1 -2 2 T -2 0 0 1 0 3 A -3 -1 -1 0 2 Case 1: xi aligns to yi Case 2: xi aligns to gap Case 3: yi aligns to gap s(xi, yj ) = +1, match -1, mismatch d=1 F(i-1, j-1)+s(xi, yj) F(i-1, j)-d F(i, j-1)-d F(0, 0) = 0 F(0, i) = −i * d F(j, 0) = −j* d PTR = DIAG, if case 1 UP, if case 2 LEFT, if case 3
Multiple Sequence Alignment § A multiple sequence alignment is a sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. § The input is a set of query sequences that are assumed to have an evolutionary relationship by which they share a lineage and are descended from a common ancestor. § From the resulting multiple sequence alignment , phylogenetic analysis can be conducted to assess the sequences shared evolutionary origins.

MSA Approaches § Dynamic programming § Progressive alignment § Iterative approach

Dynamic Programming § § Direct method for MSA to identify the globally optimal alignment solution. Computational complexity § n-dimensional equivalent of the pairwise alignment matrix is formed. § The search space increases exponentially with increasing n and is strongly dependent on sequence length(N). § O(Nn)
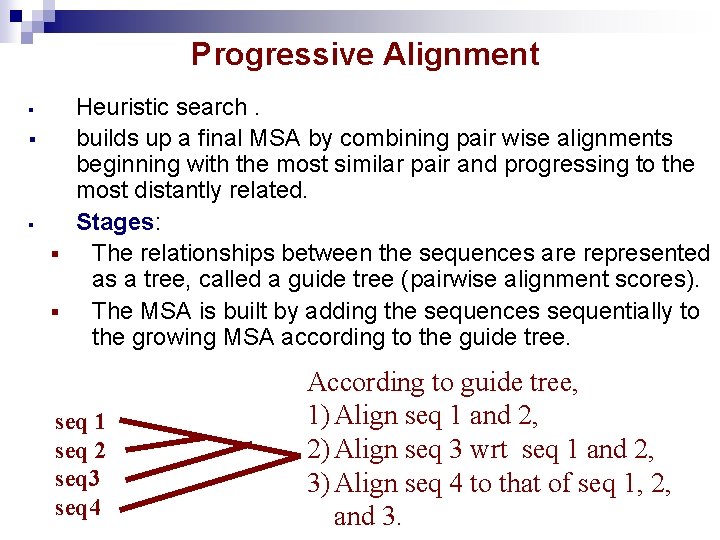
Progressive Alignment § § § Heuristic search. builds up a final MSA by combining pair wise alignments beginning with the most similar pair and progressing to the most distantly related. Stages: § The relationships between the sequences are represented as a tree, called a guide tree (pairwise alignment scores). § The MSA is built by adding the sequences sequentially to the growing MSA according to the guide tree. seq 1 seq 2 seq 3 seq 4 According to guide tree, 1) Align seq 1 and 2, 2) Align seq 3 wrt seq 1 and 2, 3) Align seq 4 to that of seq 1, 2, and 3.

Drawbacks § The primary problem is that when errors are made at any stage in growing the MSA, these errors are then propagated through to the final result. Random/ iterative approaches are used § Performance is also particularly bad when all of the sequences in the set are rather distantly related.
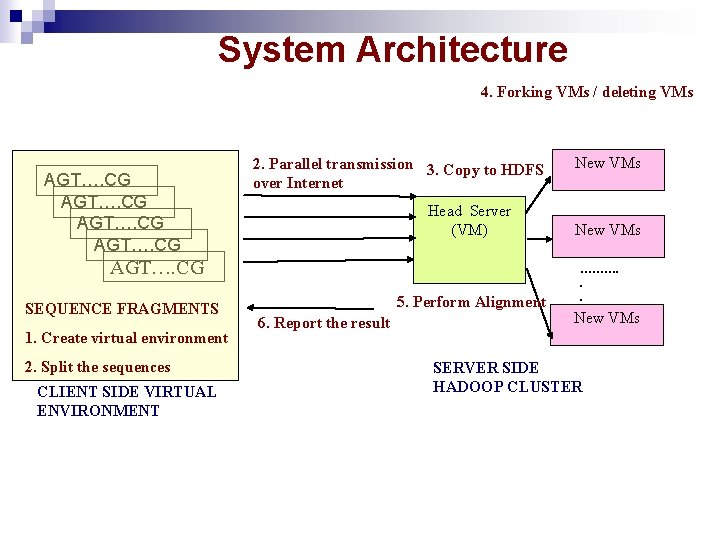
System Architecture 4. Forking VMs / deleting VMs AGT…. CG 2. Parallel transmission 3. Copy to HDFS over Internet Head Server (VM) AGT…. CG SEQUENCE FRAGMENTS 1. Create virtual environment 2. Split the sequences CLIENT SIDE VIRTUAL ENVIRONMENT 5. Perform Alignment 6. Report the result New VMs ………. . . New VMs SERVER SIDE HADOOP CLUSTER
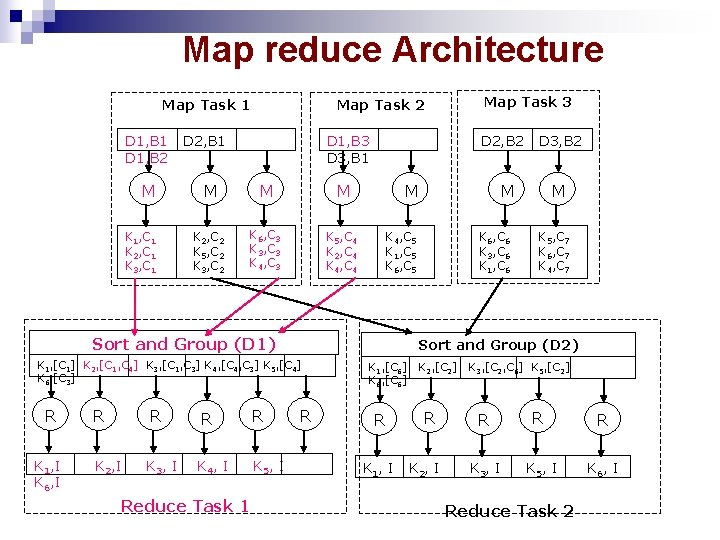
Map reduce Architecture Map Task 1 D 1, B 2 M K 1, C 1 K 2, C 1 K 3, C 1 D 2, B 1 D 1, B 3 D 3, B 1 M M M K 2, C 2 K 5, C 2 K 3, C 2 K 6, C 3 K 3, C 3 K 4, C 3 K 5, C 4 K 2, C 4 K 4, C 4 D 2, B 2 M K 1, I K 6, I R R R K 2, I K 3, I K 4, I K 5, I Reduce Task 1 K 6, C 6 K 3, C 6 K 1, C 6 M K 5, C 7 K 6, C 7 K 4, C 7 Sort and Group (D 2) K 1, [C 1] K 2, [C 1, C 4] K 3, [C 1, C 3] K 4, [C 4, C 3] K 5, [C 4] K 6, [C 3] R D 3, B 2 M K 4, C 5 K 1, C 5 K 6, C 5 Sort and Group (D 1) R Map Task 3 Map Task 2 R K 1, [C 6] K 6, [C 6] R K 1, I K 2, [C 2] R K 2, I K 3, [C 2, C 6] K 5, [C 2] R R R K 3, I K 5, I K 6, I Reduce Task 2
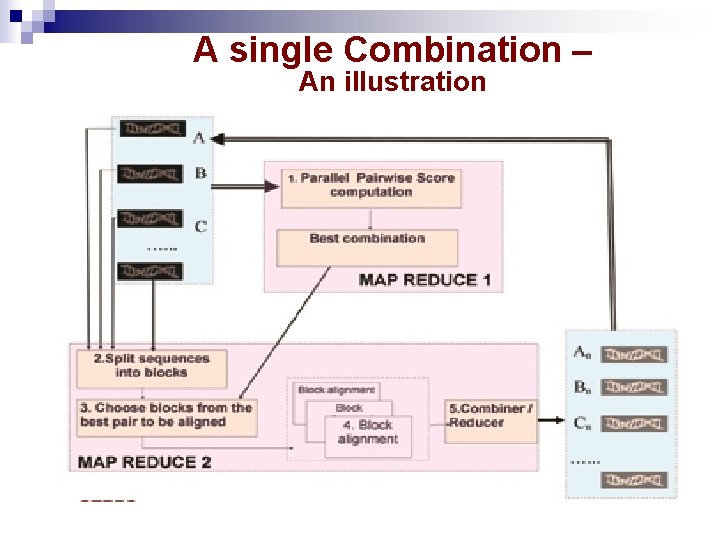
A single Combination – An illustration
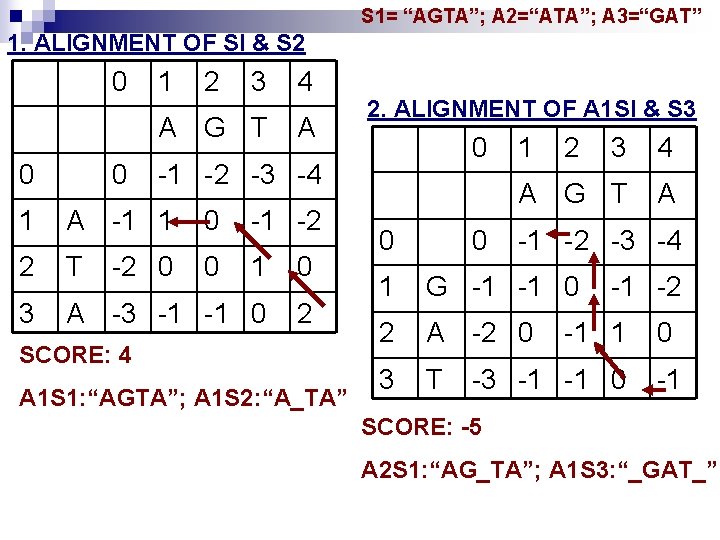
S 1= “AGTA”; A 2=“ATA”; A 3=“GAT” 1. ALIGNMENT OF SI & S 2 0 1 2 3 4 A G T A 0 0 1 A -1 1 0 -1 -2 2 T 0 1 0 3 A -3 -1 -1 0 2 2. ALIGNMENT OF A 1 SI & S 3 0 -1 -2 -3 -4 -2 0 SCORE: 4 A 1 S 1: “AGTA”; A 1 S 2: “A_TA” 0 0 1 2 3 4 A G T A -1 -2 -3 -4 1 G -1 -1 0 2 A -2 0 3 T -1 -2 -1 1 -3 -1 -1 0 0 -1 SCORE: -5 A 2 S 1: “AG_TA”; A 1 S 3: “_GAT_”
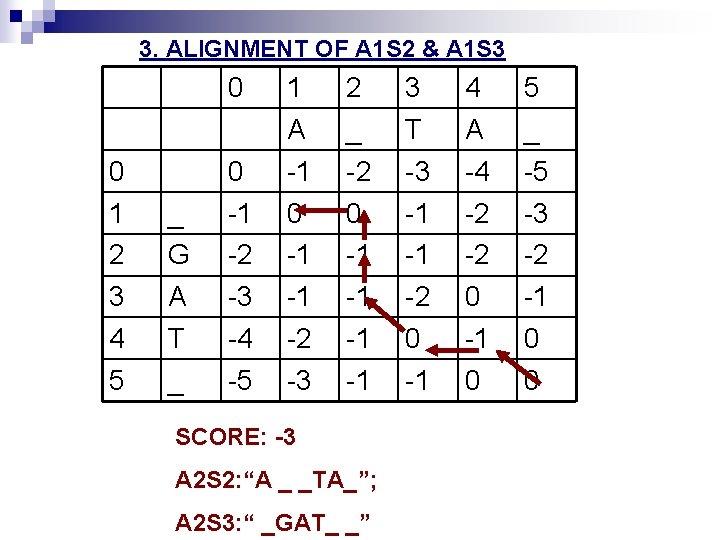
3. ALIGNMENT OF A 1 S 2 & A 1 S 3 0 0 1 2 3 4 5 _ G A T _ 0 -1 -2 -3 -4 -5 1 A -1 0 -1 -1 -2 -3 2 _ -2 0 -1 -1 SCORE: -3 A 2 S 2: “A _ _TA_”; A 2 S 3: “ _GAT_ _” 3 T -3 -1 -1 -2 0 -1 4 A -4 -2 -2 0 -1 0 5 _ -5 -3 -2 -1 0 0
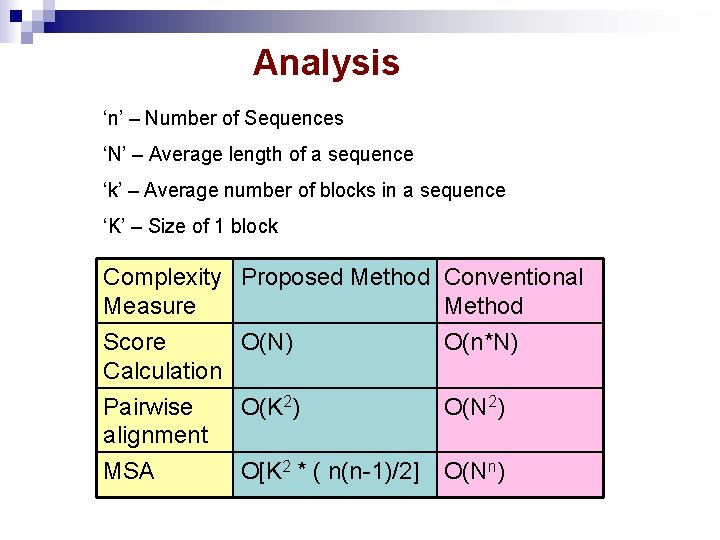
Analysis ‘n’ – Number of Sequences ‘N’ – Average length of a sequence ‘k’ – Average number of blocks in a sequence ‘K’ – Size of 1 block Complexity Proposed Method Conventional Measure Method Score O(N) Calculation Pairwise O(K 2) alignment O(n*N) MSA O(Nn) O[K 2 * ( n(n-1)/2] O(N 2)
2. Parallelised data trasfer ‘T’ – Time for sequence transfer serially & ‘k’ – block size T/k – Time for sequence transfer in parallel 3. Dynamic cluster creation Advantage: Computation power of remote cluster is optimal and not wasted Disadvantage: Time to set up the cluster
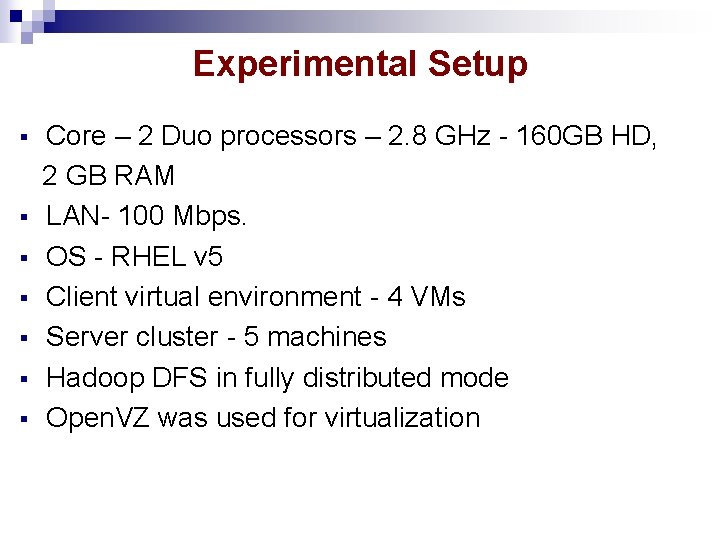
Experimental Setup § § § § Core – 2 Duo processors – 2. 8 GHz - 160 GB HD, 2 GB RAM LAN- 100 Mbps. OS - RHEL v 5 Client virtual environment - 4 VMs Server cluster - 5 machines Hadoop DFS in fully distributed mode Open. VZ was used for virtualization
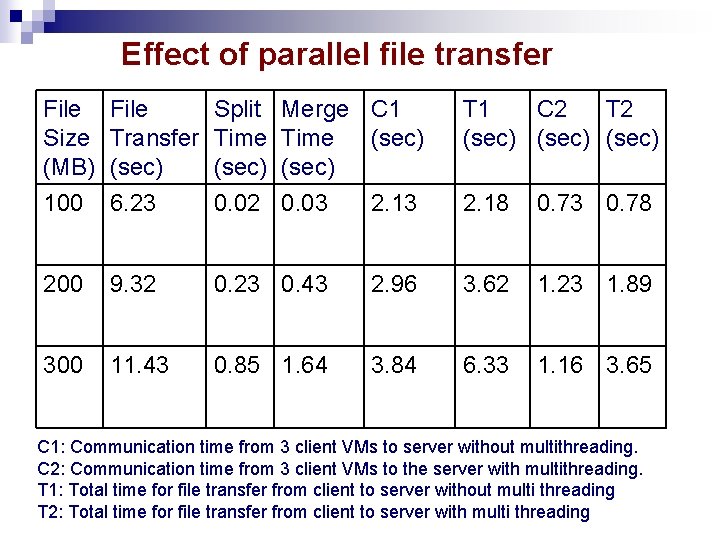
Effect of parallel file transfer File Size (MB) 100 File Transfer (sec) 6. 23 Split Time (sec) 0. 02 Merge C 1 Time (sec) 0. 03 2. 13 200 9. 32 0. 23 0. 43 300 11. 43 0. 85 1. 64 T 1 C 2 T 2 (sec) 2. 18 0. 73 0. 78 2. 96 3. 62 1. 23 1. 89 3. 84 6. 33 1. 16 3. 65 C 1: Communication time from 3 client VMs to server without multithreading. C 2: Communication time from 3 client VMs to the server with multithreading. T 1: Total time for file transfer from client to server without multi threading T 2: Total time for file transfer from client to server with multi threading
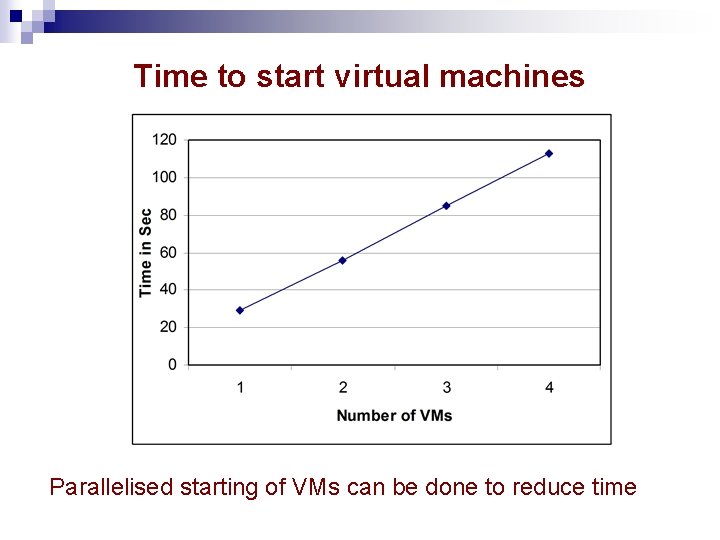
Time to start virtual machines Parallelised starting of VMs can be done to reduce time
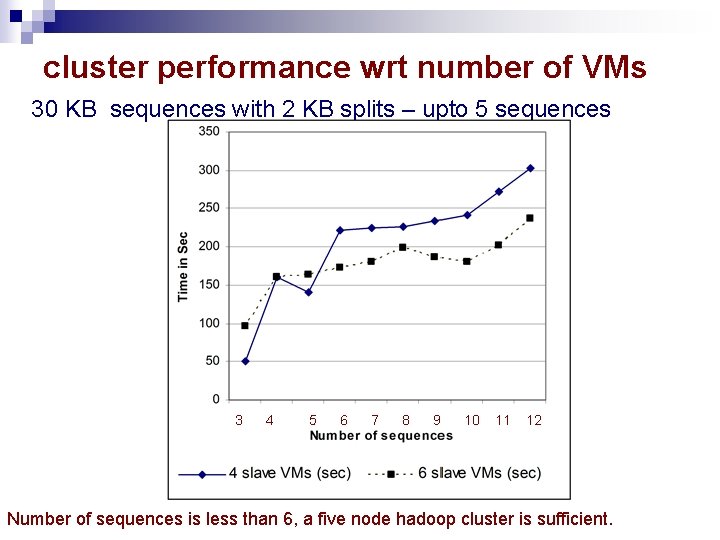
cluster performance wrt number of VMs 30 KB sequences with 2 KB splits – upto 5 sequences 3 4 5 6 7 8 9 10 11 12 Number of sequences is less than 6, a five node hadoop cluster is sufficient.
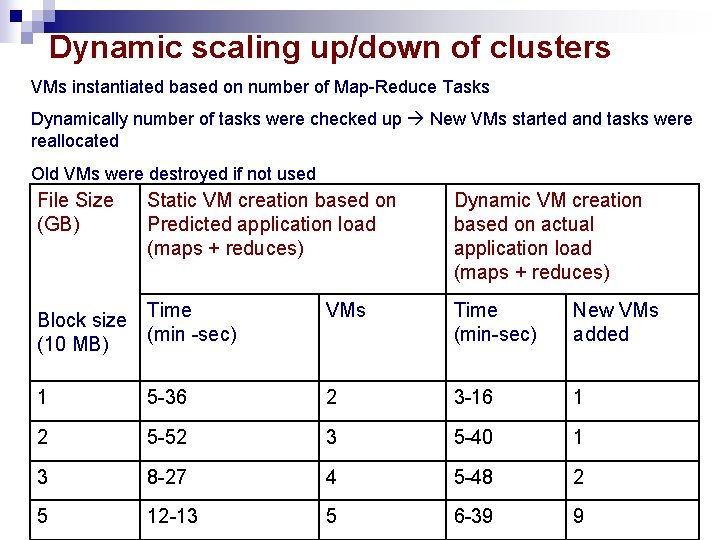
Dynamic scaling up/down of clusters VMs instantiated based on number of Map-Reduce Tasks Dynamically number of tasks were checked up New VMs started and tasks were reallocated Old VMs were destroyed if not used File Size (GB) Static VM creation based on Predicted application load (maps + reduces) Dynamic VM creation based on actual application load (maps + reduces) Block size (10 MB) Time (min -sec) VMs Time (min-sec) New VMs added 1 5 -36 2 3 -16 1 2 5 -52 3 5 -40 1 3 8 -27 4 5 -48 2 5 12 -13 5 6 -39 9
Conclusion 1) Proposed MSA improves on the computation time and also maintains the accuracy. § Parallelism of sequence alignment in three levels. Hadoop data grids - Data and compute parallelism & scalability § Dynamic Programming - accuracy. 2) Complexity is reduced from O(Nn) to O[K 2 * (n *(n-1)/2)] n Combining progressive and dynamic approaches. n Blocking in hadoop 3) Enhancements (using clouds for MSA) n Automatic configuration of the cloud environment based on the computational needs n Efficient upload of data into the HDFS by parallel transfer of sequence fragments over the Internet.

Acknowledgements The Research has been carried out as a result of PSG-Yahoo Research programme on Grid and Cloud computing. Sincere Thanks to 1) Dr R Rudramoorthy, Principal, PSG College of Techniology, Coimbatore. 2) Mr K V Chidambaran, Director, Grid and Cloud Systems Group, Yahoo, Bangalore

THANK YOU QUESTIONS?

REFERENCES n Apache, (2002), Hadoop Documentation, retrieved on September 20, 2009, fromhttp: //hadoop. apache. org/core/docs/r 0. 17. 2/. n Tahir, N. , Imitaz, S. and Shaftab, A. , “Parallel Needleman-Wunsch Algorithm for Grid”. retrieved on January 19, 2009 from http: //www. gridbus. org/~alchemi/files/Parallel%20 Needleman% 20 Algo. pdf n Michael, C. , (2009). “Cloud Burst: highly sensitive read mapping with Map. Reduce”, Bioinformatics, 25(11), 1363 -1369. n Lee, T. , “A genomic Clu. E for Cloud Computing”, retrieved on January 13, 2009 from http: //www. eurekalert. org/pub_releases /2009 -04/uomagc 042309. php n Yongli, H. and Shen, J. , “Sequence analysis scale up and acceleration using Grid and Cloud Computing yield efficient analyses of HIV-1 variants and other viruses”, retrieved on February 15, 2009 from www. iscb. org /uploaded/css/43/12056. pdf. n Philip, P. , Andres, L. , Eyal, L. and Michael, B. “Adding the easy button to the cloud with Snow. Flock and MPI”, in Proceedings of 3 rd ACM workshop in system level virtualization for HPC (2009), 122 -127.
- Slides: 31