Parallel Medical and Genomics Applications on Power 3




- Slides: 49

Parallel Medical and Genomics Applications on Power 3 and Power 4 Machines Amit Majumdar San Diego Supercomputer Center - UCSD Application I : Brain Deformation Simulation in Image Guided Neurosurgery Simon K. Warfield 1, Florin Talos 1, 2, Alida Tei 1, 3, Aditya Bharatha 1, 4, Arya Nabavi 1, 2, Matthieu Ferrante 1, 5, Peter Mc. L. Black 2, Ferenc A. Jolesz 1, Ron Kikinis 1, Corey Kemper 1 1 Surgical Planing Laboratory and 2 Dept. of Surgery Brigham and Women’s Hospital and Harvard Medical School 3 Massashusetts Institute of Technology 4 University of Toronto Medical School 5 Telecom. Lab. , Universite’ Catholique de Louvain, Belgium Application II : Monte Carlo SPECT Imaging Yuni Dewaraja 1, Kenneth Koral 1, Abhijit Bose 2, Michael Ljungberg 3 1 Nuclear Medicine, University of Michigan 2 Center for Advanced Computing, University of Michigan 3 Department of Radiation Physics, Univ. of Lund, Sweden Application III : Parallel Proteomics Application John R. Yates 1, Daniel J. Carucci 2 , Giri Chukkapalli 3, Robert Sinkovits 3 1 The Scripps Research Institute 2 Naval Medical Research Center, US NAVY 3 SDSC SP Sci. Comp 6, Berkeley, August 23, 2002

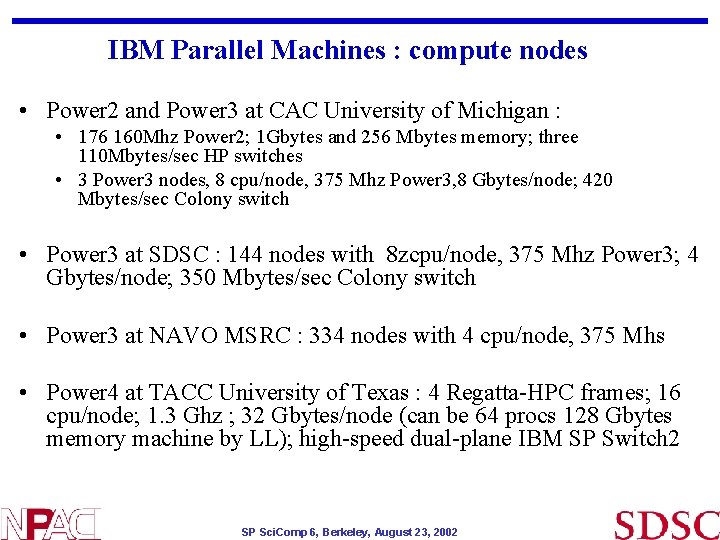
IBM Parallel Machines : compute nodes • Power 2 and Power 3 at CAC University of Michigan : • 176 160 Mhz Power 2; 1 Gbytes and 256 Mbytes memory; three 110 Mbytes/sec HP switches • 3 Power 3 nodes, 8 cpu/node, 375 Mhz Power 3, 8 Gbytes/node; 420 Mbytes/sec Colony switch • Power 3 at SDSC : 144 nodes with 8 zcpu/node, 375 Mhz Power 3; 4 Gbytes/node; 350 Mbytes/sec Colony switch • Power 3 at NAVO MSRC : 334 nodes with 4 cpu/node, 375 Mhs • Power 4 at TACC University of Texas : 4 Regatta-HPC frames; 16 cpu/node; 1. 3 Ghz ; 32 Gbytes/node (can be 64 procs 128 Gbytes memory machine by LL); high-speed dual-plane IBM SP Switch 2 SP Sci. Comp 6, Berkeley, August 23, 2002

Application I : Brain Deformation Simulation in Image Guided Neurosurgery SP Sci. Comp 6, Berkeley, August 23, 2002

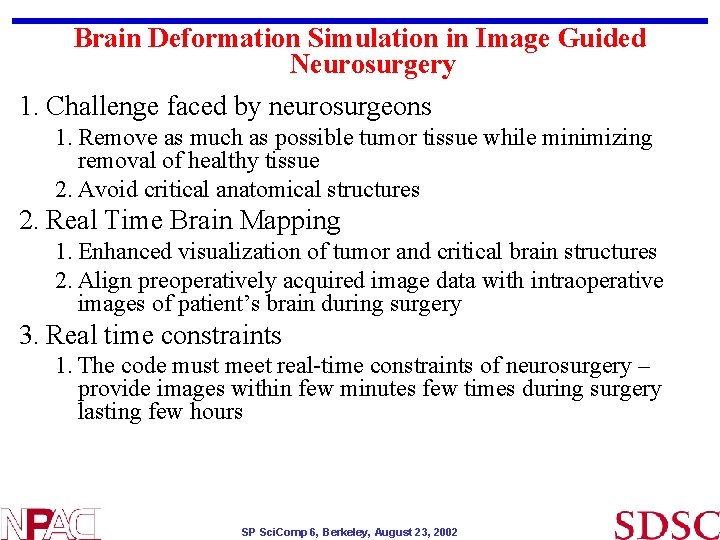
Brain Deformation Simulation in Image Guided Neurosurgery 1. Challenge faced by neurosurgeons 1. Remove as much as possible tumor tissue while minimizing removal of healthy tissue 2. Avoid critical anatomical structures 2. Real Time Brain Mapping 1. Enhanced visualization of tumor and critical brain structures 2. Align preoperatively acquired image data with intraoperative images of patient’s brain during surgery 3. Real time constraints 1. The code must meet real-time constraints of neurosurgery – provide images within few minutes few times during surgery lasting few hours SP Sci. Comp 6, Berkeley, August 23, 2002

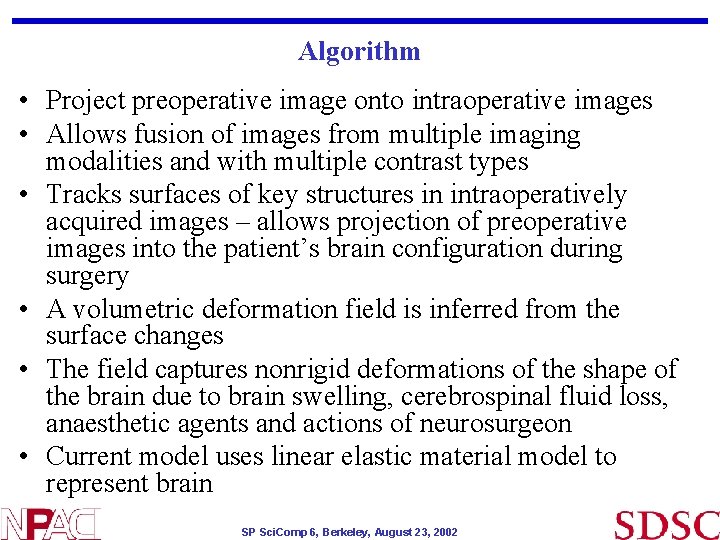
Algorithm • Project preoperative image onto intraoperative images • Allows fusion of images from multiple imaging modalities and with multiple contrast types • Tracks surfaces of key structures in intraoperatively acquired images – allows projection of preoperative images into the patient’s brain configuration during surgery • A volumetric deformation field is inferred from the surface changes • The field captures nonrigid deformations of the shape of the brain due to brain swelling, cerebrospinal fluid loss, anaesthetic agents and actions of neurosurgeon • Current model uses linear elastic material model to represent brain SP Sci. Comp 6, Berkeley, August 23, 2002

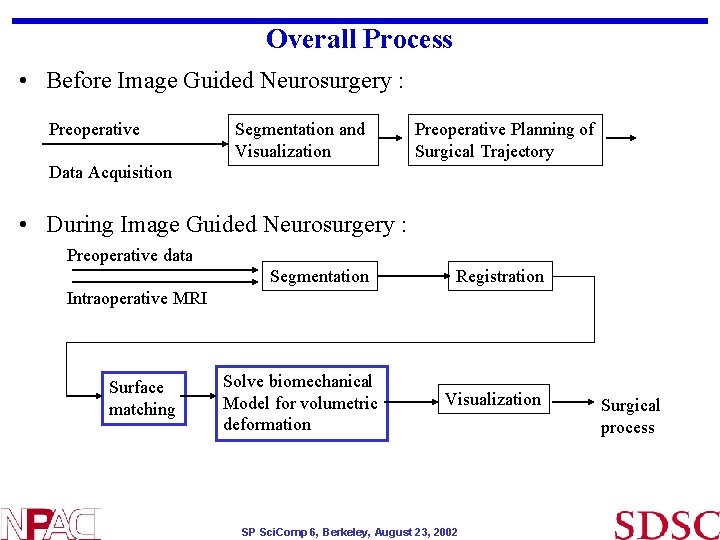
Overall Process • Before Image Guided Neurosurgery : Preoperative Segmentation and Visualization Preoperative Planning of Surgical Trajectory Data Acquisition • During Image Guided Neurosurgery : Preoperative data Segmentation Registration Intraoperative MRI Surface matching Solve biomechanical Model for volumetric deformation Visualization SP Sci. Comp 6, Berkeley, August 23, 2002 Surgical process

Volumetric Biomechanical Simulation of Brain Deformation • During surgery brain shape changes due to surgical intervention • During surgery surgeon can acquire new volumetric MRI to review current configuration of the entire brain • Volumetric Biomechanical Simulation of Brain Deformation • Match surface from earlier acquisition to the new acquisition • Infer volumetric deformation based upon the surface correspondences • Apply forces to the volumetric model that will produce the same displacement field at the surface as was obtained by the surface matching • Biomechanical model allows the computation of the deformation throughout the volume SP Sci. Comp 6, Berkeley, August 23, 2002

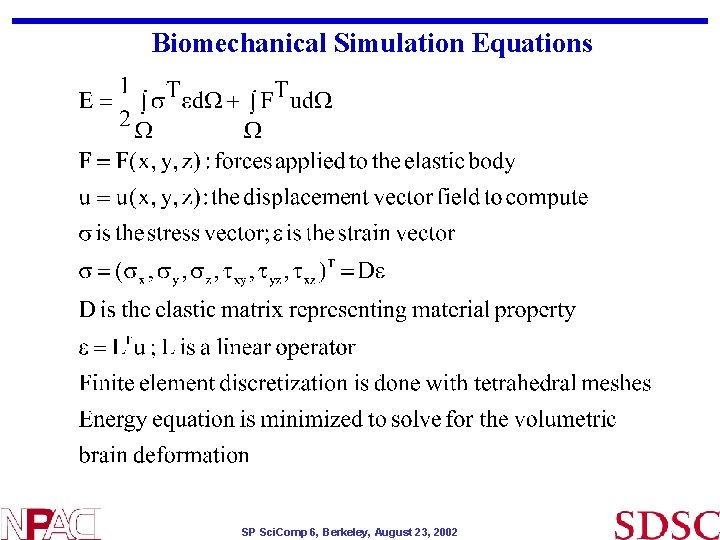
Biomechanical Simulation Equations SP Sci. Comp 6, Berkeley, August 23, 2002

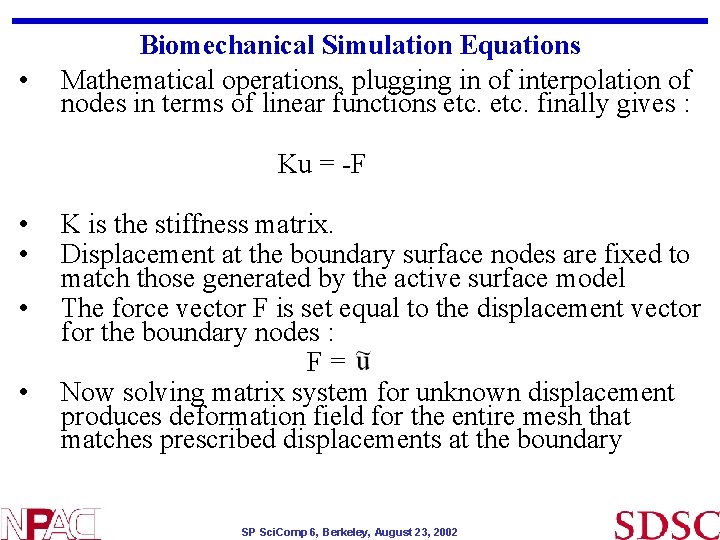
• Biomechanical Simulation Equations Mathematical operations, plugging in of interpolation of nodes in terms of linear functions etc. finally gives : Ku = -F • • K is the stiffness matrix. Displacement at the boundary surface nodes are fixed to match those generated by the active surface model The force vector F is set equal to the displacement vector for the boundary nodes : F= Now solving matrix system for unknown displacement produces deformation field for the entire mesh that matches prescribed displacements at the boundary SP Sci. Comp 6, Berkeley, August 23, 2002

Signa SP (GE Medical Systems) R. Pergolizzi SP Sci. Comp 6, Berkeley, August 23, 2002

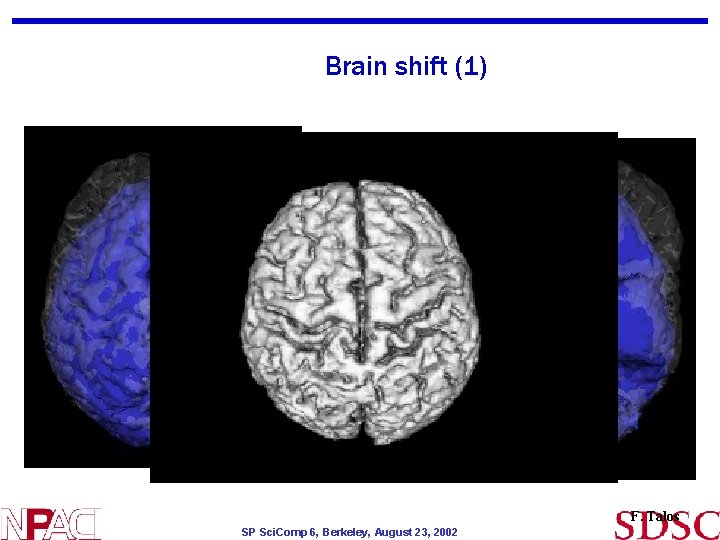
Brain shift (1) F. Talos SP Sci. Comp 6, Berkeley, August 23, 2002

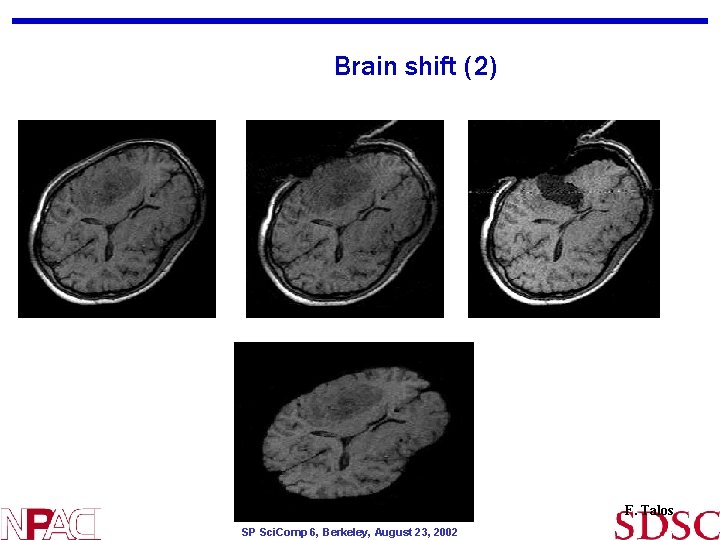
Brain shift (2) F. Talos SP Sci. Comp 6, Berkeley, August 23, 2002

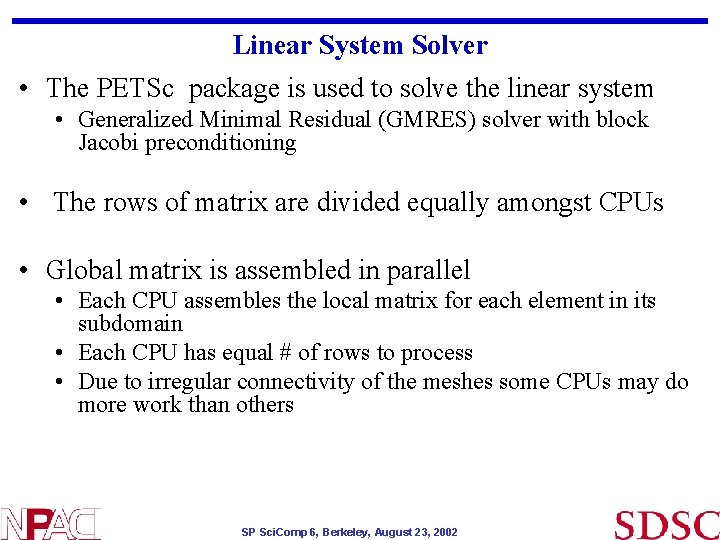
Linear System Solver • The PETSc package is used to solve the linear system • Generalized Minimal Residual (GMRES) solver with block Jacobi preconditioning • The rows of matrix are divided equally amongst CPUs • Global matrix is assembled in parallel • Each CPU assembles the local matrix for each element in its subdomain • Each CPU has equal # of rows to process • Due to irregular connectivity of the meshes some CPUs may do more work than others SP Sci. Comp 6, Berkeley, August 23, 2002

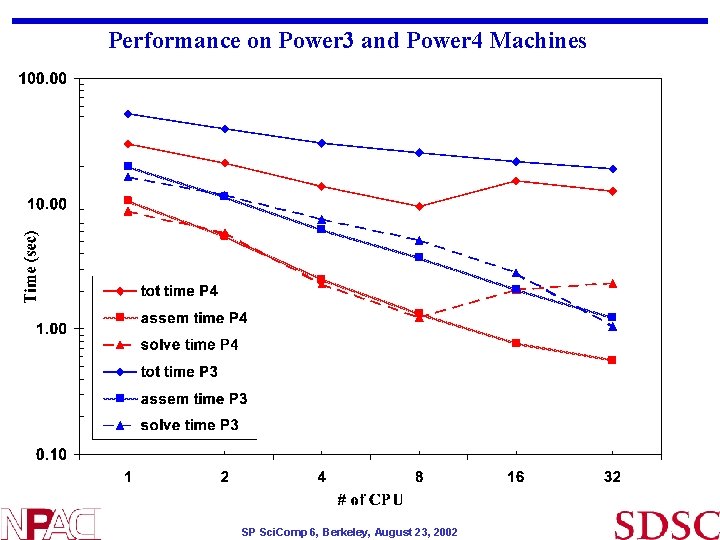
Performance on Power 3 and Power 4 Machines SP Sci. Comp 6, Berkeley, August 23, 2002

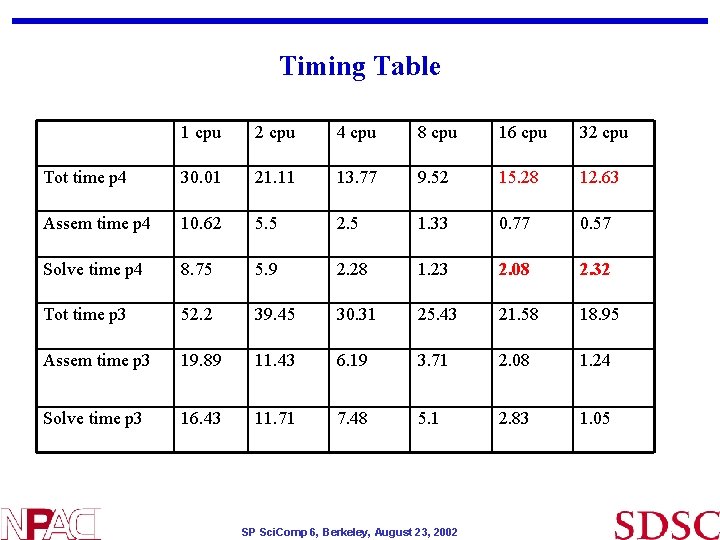
Timing Table 1 cpu 2 cpu 4 cpu 8 cpu 16 cpu 32 cpu Tot time p 4 30. 01 21. 11 13. 77 9. 52 15. 28 12. 63 Assem time p 4 10. 62 5. 5 2. 5 1. 33 0. 77 0. 57 Solve time p 4 8. 75 5. 9 2. 28 1. 23 2. 08 2. 32 Tot time p 3 52. 2 39. 45 30. 31 25. 43 21. 58 18. 95 Assem time p 3 19. 89 11. 43 6. 19 3. 71 2. 08 1. 24 Solve time p 3 16. 43 11. 71 7. 48 5. 1 2. 83 1. 05 SP Sci. Comp 6, Berkeley, August 23, 2002

Observations • Power 4 timings: • Unexpected timings on Power 4 16 and 32 processors • Scheduler gives exclusive access to nodes and CPUs • Cache , network ? • Power 3 timing is consistent (with other machines) • Overall scaling is not good beyond few processors • Serial I/O part contributes to this • Petsc performance : (GMRES with block jacobi precond) • Linear system solver MFLOPS scale well with # of procs on Power 3 (have not checked on Power 4 yet) • Petsc sparse matrix storage allocation is efficient • # of GMRES iterations increase with # of processors – 41 to 135 iterations on 1 to 16 procs respectively - contributes partially to scaling • Future plan is to investigate scaling further • Investigate viscoelastic modelling ($funding$) • Thanks to Petsc group (Barry Smith) for valuable discussions SP Sci. Comp 6, Berkeley, August 23, 2002

Application II : Monte Carlo SPECT Imaging SP Sci. Comp 6, Berkeley, August 23, 2002

• Radionuclide therapy • SPECT imaging in radionuclide therapy • Monte Carlo simulation of SPECT imaging • SIMIND code • Applications • Parallel Monte Carlo code and performance SP Sci. Comp 6, Berkeley, August 23, 2002

Radionuclide therapy • Cancer cells are sterilized using internally administered ionizing radiation • Some therapeutic isotopes, ex. I-131, produce both beta particles and gamma ray photons • Beta particles kill tumor cells. Beta pathlength span several cells. • Photons used to image radioactivity distribution within patient • Radionuclide therapy has less toxic effect on normal tissue than chemotherapy. SP Sci. Comp 6, Berkeley, August 23, 2002

I-131 Radionuclide therapy • I-131 Radioimmunotherapy (RIT) : I-131 labeled antibodies selectively target radioactivity to tumor cells while sparing normal tissue. • Shows promise for the treatment of non-Hodgkin’s lymphoma (NHL). NHL is the fifth leading cause of cancer death. Median survival 6 -10 years. • I-131 MIBG • Shows promise for the treatment of metastatic neuroblastoma which is a childhood cancer with poor long term survival. SP Sci. Comp 6, Berkeley, August 23, 2002

I-131 RIT for NHL at University of Michigan • Phase II clinical trial: Out of 76 patients with no previous treatment, 48 achieved a complete response and 26 achieved a partial response. • Patient-specific infusion • Tracer dose: ~ 5 m. Ci for dosimetry studies to determine therapeutic dose for each patient • Therapeutic dose: 50 -100 m. Ci one week later SP Sci. Comp 6, Berkeley, August 23, 2002

I-131 imaging • Single photon emission computed tomography (SPECT) imaging using a rotating gamma camera • Components of the gamma camera • Lead collimator • Detection medium - scintillation crystal • Electronics • Tomographic reconstruction of SPECT data produces a 3 -D image of the radioactivity distribution within the patient. SP Sci. Comp 6, Berkeley, August 23, 2002

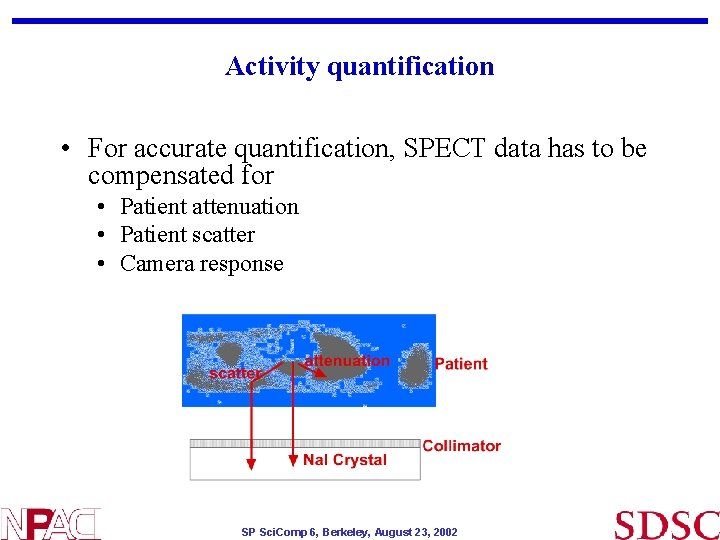
Activity quantification • For accurate quantification, SPECT data has to be compensated for • Patient attenuation • Patient scatter • Camera response SP Sci. Comp 6, Berkeley, August 23, 2002

What is the role of M. C. simulation in SPECT Imaging? • Monte Carlo is used primarily to evaluate compensation methods for scatter, attenuation and camera response and to evaluate the overall accuracy of our clinical activity quantification. • M. C. is ideal for such evaluations because unlike in experiments, the details of photon histories are known. SP Sci. Comp 6, Berkeley, August 23, 2002

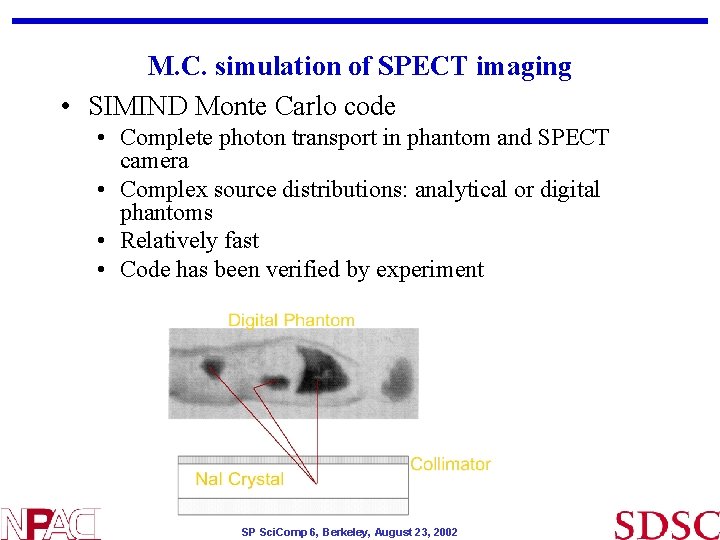
M. C. simulation of SPECT imaging • SIMIND Monte Carlo code • Complete photon transport in phantom and SPECT camera • Complex source distributions: analytical or digital phantoms • Relatively fast • Code has been verified by experiment SP Sci. Comp 6, Berkeley, August 23, 2002

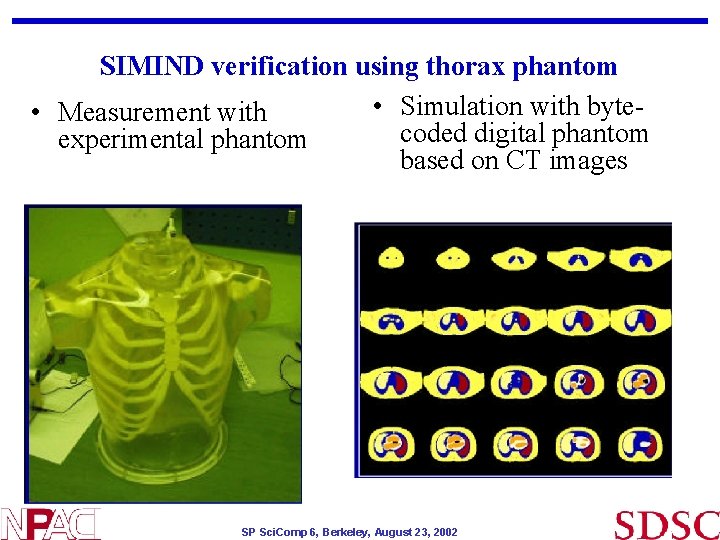
SIMIND verification using thorax phantom • Simulation with byte • Measurement with coded digital phantom experimental phantom based on CT images SP Sci. Comp 6, Berkeley, August 23, 2002

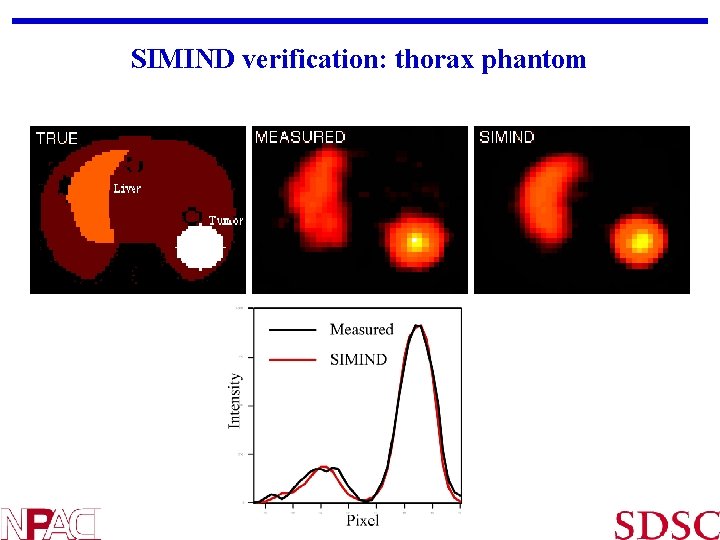
SIMIND verification: thorax phantom SP Sci. Comp 6, Berkeley, August 23, 2002

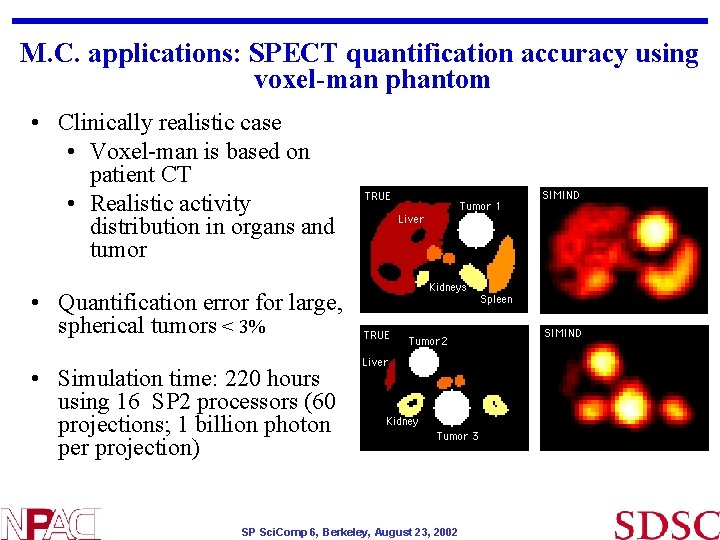
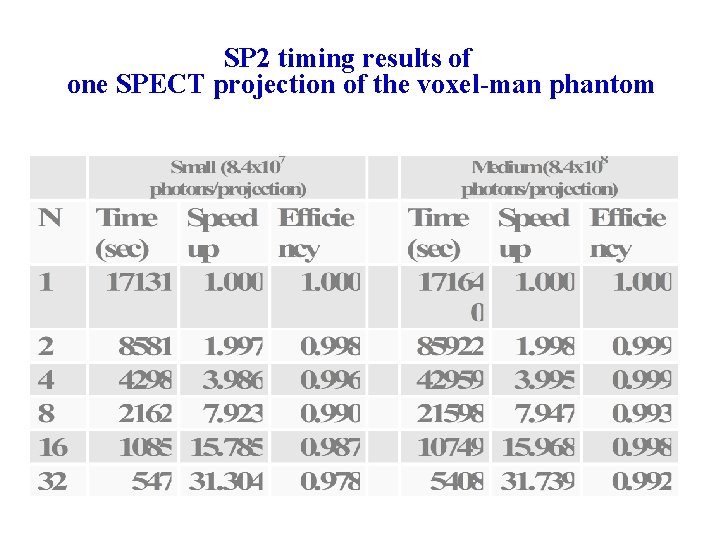
M. C. applications: SPECT quantification accuracy using voxel-man phantom • Clinically realistic case • Voxel-man is based on patient CT • Realistic activity distribution in organs and tumor • Quantification error for large, spherical tumors < 3% • Simulation time: 220 hours using 16 SP 2 processors (60 projections; 1 billion photon per projection) SP Sci. Comp 6, Berkeley, August 23, 2002

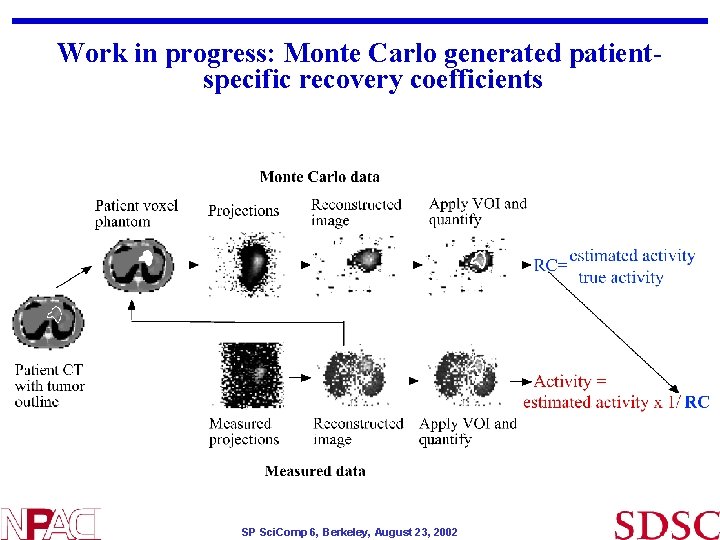
Work in progress: Monte Carlo generated patientspecific recovery coefficients SP Sci. Comp 6, Berkeley, August 23, 2002

A parallel M. C. code for SPECT: motivation • Fast code for accurate I-131 Monte Carlo simulations • I-131 M. C. simulations are computationally tedious • Physical modeling of collimator • Variance reduction limited • SPECT require a large number of projections • Realistic simulations using high resolution voxel phantoms • When all of the above are included in a simulation, CPU time can be several months using the serial SIMIND code SP Sci. Comp 6, Berkeley, August 23, 2002

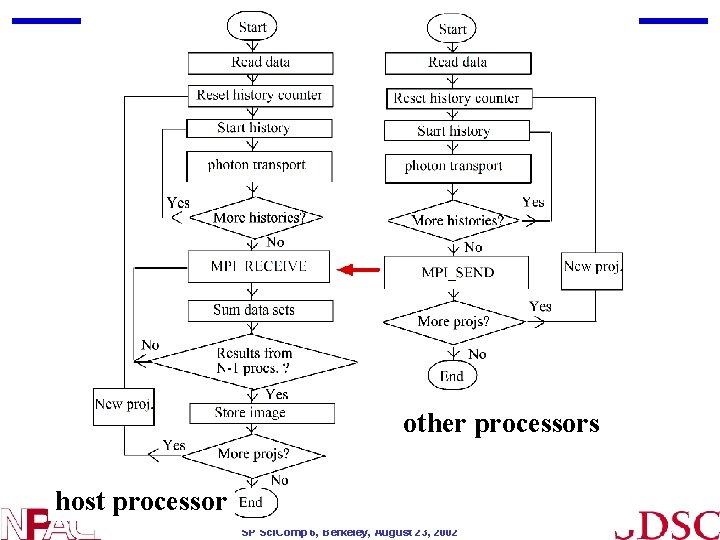
SIMIND parallelization • In the present application the photon histories are independent of each other - “inherently parallel” • Critical to have a good parallel RNG (SPRNG) • Each processor performs entire simulation and reports results to host processor • Code replicated in each of the N processors • Host sums N partial results and calculates the final result • Standard deviation of the combined result is improved by 1/sqrt(N) • Minimal changes to original SIMIND code SP Sci. Comp 6, Berkeley, August 23, 2002

other processors host processor SP Sci. Comp 6, Berkeley, August 23, 2002

SP 2 timing results of one SPECT projection of the voxel-man phantom

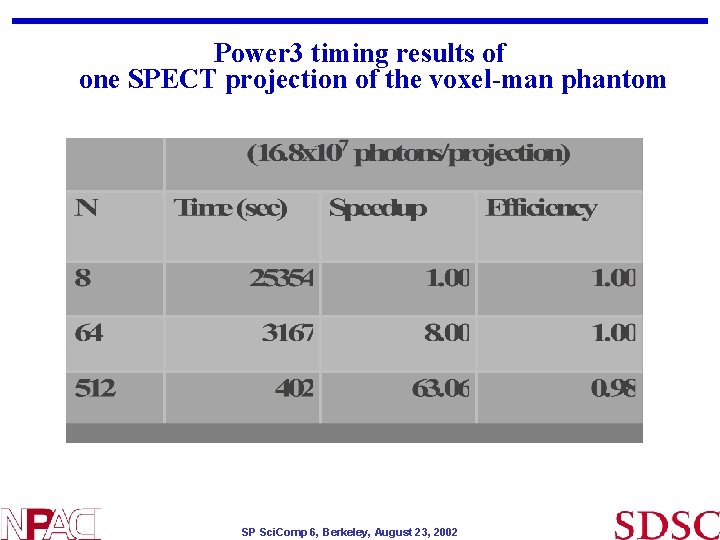
Power 3 timing results of one SPECT projection of the voxel-man phantom SP Sci. Comp 6, Berkeley, August 23, 2002

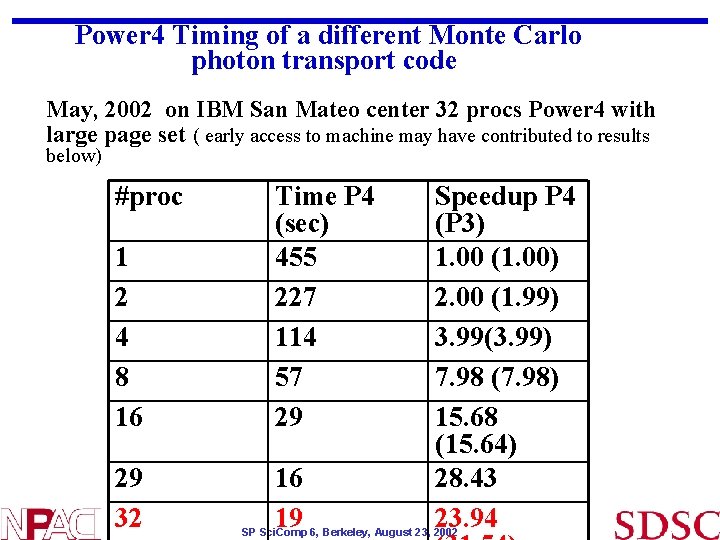
Power 4 Timing of a different Monte Carlo photon transport code May, 2002 on IBM San Mateo center 32 procs Power 4 with large page set ( early access to machine may have contributed to results below) #proc 1 2 4 8 16 Time P 4 (sec) 455 227 114 57 29 29 32 16 19 Speedup P 4 (P 3) 1. 00 (1. 00) 2. 00 (1. 99) 3. 99(3. 99) 7. 98 (7. 98) 15. 68 (15. 64) 28. 43 23. 94 SP Sci. Comp 6, Berkeley, August 23, 2002

More Realistic Simulations • A 60 -projection SPECT simulation of the voxel-man phantom simulation of the voxel-phantom. • Realistic values were used for the activity concentration ratios in several organs and tumors (based on typical I-131 RIT patient studies at U. Michigan clinic): kidneys, 81; liver, 26; lungs, 26; spleen 53; blood pool, 48; 100 cc spherical tumor, 100; 50 cc spherical tumor, 100; 20 cc spherical tumor, 100; all other structures, 4. The SPECT matrix size was 64 x 64 with pixel size of 0. 8 cm x 0. 4 cm. Up to 3 orders of scatter was modeled. 1 billion photons were simulated for each projection. • The simulation time on Power 3 using 512 processors was 6. 5 hours for all 60 projections (time for each projection was 6. 5 min). SP Sci. Comp 6, Berkeley, August 23, 2002

Effect of Parallel Computing • Monte Carlo has enabled to evaluate and improve the quantification of I-131 tumor uptake for dosimetry in NHL patients undergoing RIT at U. Michigan clinic. • May lead to statistically significant dose-response relationships • Speed-up due to the parallel SIMIND code has enabled us to carry out clinically realistic simulations using voxel-phantoms. • In the future we will carry out M. C. based dose calculations • More accurate • Tumor and organ dose distributions SP Sci. Comp 6, Berkeley, August 23, 2002

Application III : Parallel Proteomics Application SP Sci. Comp 6, Berkeley, August 23, 2002

• Sequest is a proteomics application used to analyze mass spectrometer output and match to protein database to identify proteins • The Naval Medical Research Center (NMRC) is using Sequest in the research to develop a malaria vaccine based on the expression of proteins in the various stages of the malaria parasite. • Performance of the serial Sequest is currently was a major bottleneck in malaria vaccine project • SDSC computational scientists developed a parallel version of the Sequest code to reduce simulation time significantly SP Sci. Comp 6, Berkeley, August 23, 2002

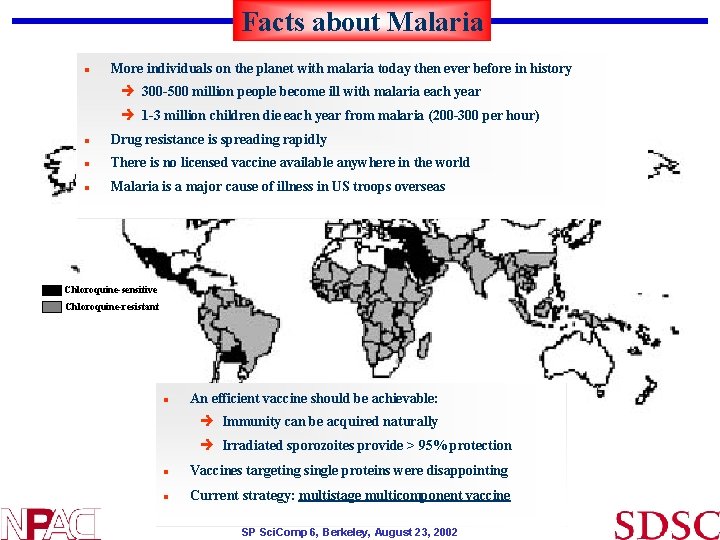
Facts about Malaria n More individuals on the planet with malaria today then ever before in history è 300 -500 million people become ill with malaria each year è 1 -3 million children die each year from malaria (200 -300 per hour) n Drug resistance is spreading rapidly n There is no licensed vaccine available anywhere in the world n Malaria is a major cause of illness in US troops overseas Chloroquine-sensitive Chloroquine-resistant n An efficient vaccine should be achievable: è Immunity can be acquired naturally è Irradiated sporozoites provide > 95% protection n Vaccines targeting single proteins were disappointing n Current strategy: multistage multicomponent vaccine SP Sci. Comp 6, Berkeley, August 23, 2002

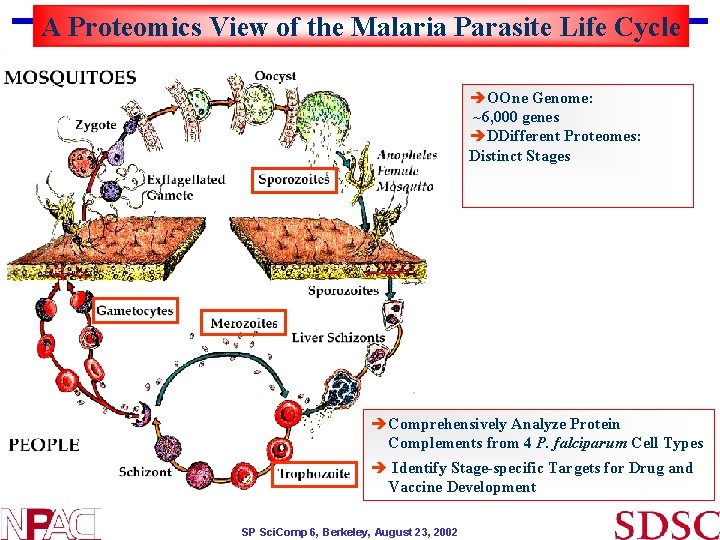
A Proteomics View of the Malaria Parasite Life Cycle èOOne Genome: ~6, 000 genes èDDifferent Proteomes: Distinct Stages èComprehensively Analyze Protein Complements from 4 P. falciparum Cell Types è Identify Stage-specific Targets for Drug and Vaccine Development SP Sci. Comp 6, Berkeley, August 23, 2002

Importance of a malaria vaccine The battle against malaria hampered by the emergence of drug resistant strains of Plasmodium falciparum, the parasite responsible for majority of malaria infections. Most cases of malaria are concentrated in the world's poorest countries. Malaria vaccine likely to be affordable alternative to expensive drugs. SP Sci. Comp 6, Berkeley, August 23, 2002

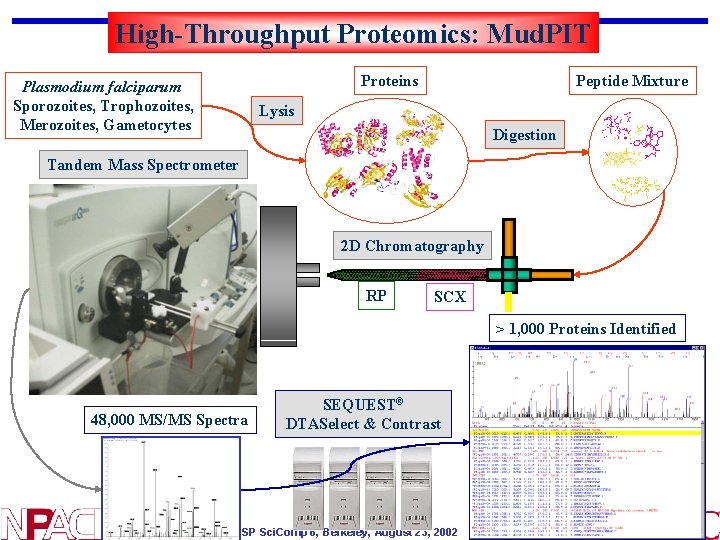
High-Throughput Proteomics: Mud. PIT Proteins Plasmodium falciparum Sporozoites, Trophozoites, Merozoites, Gametocytes Peptide Mixture Lysis Digestion Tandem Mass Spectrometer 2 D Chromatography RP SCX > 1, 000 Proteins Identified 48, 000 MS/MS Spectra SEQUEST® DTASelect & Contrast SP Sci. Comp 6, Berkeley, August 23, 2002

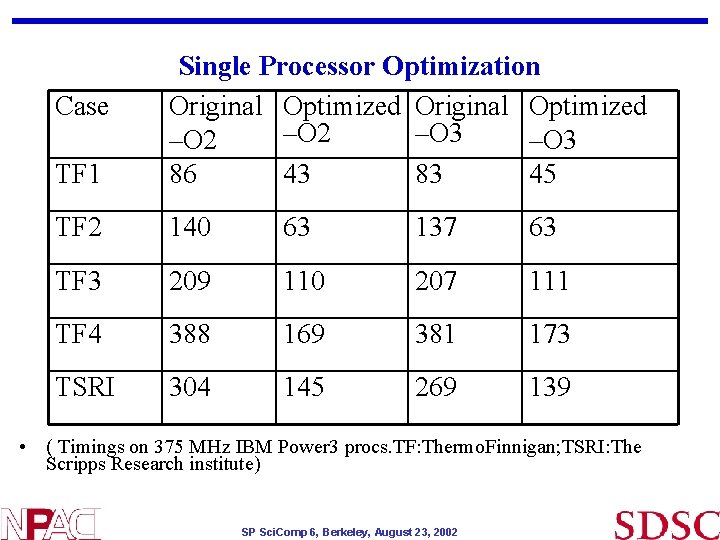
TF 1 Single Processor Optimization Original Optimized –O 2 –O 3 86 43 83 45 TF 2 140 63 137 63 TF 3 209 110 207 111 TF 4 388 169 381 173 TSRI 304 145 269 139 Case • ( Timings on 375 MHz IBM Power 3 procs. TF: Thermo. Finnigan; TSRI: The Scripps Research institute) SP Sci. Comp 6, Berkeley, August 23, 2002

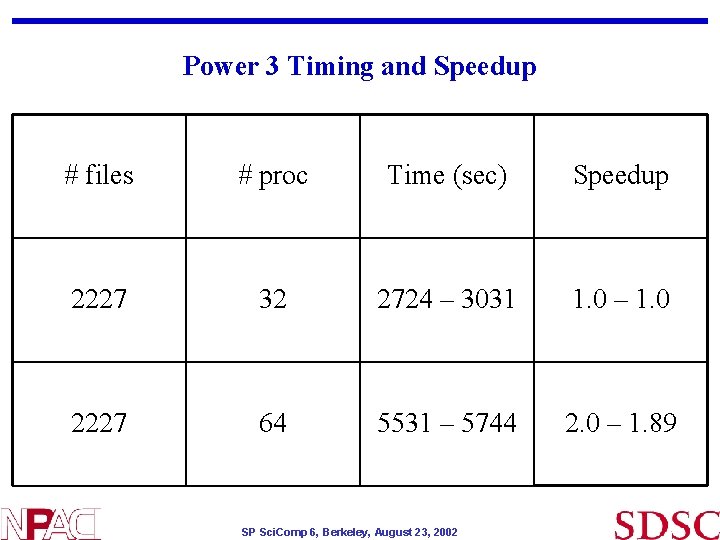
Code parallelization • Parallel version of Sequest has been developed using MPI and incorporating all single processor optimizations. • Parallelization was done so that all the processors work on a different file simultaneously; files are picked up from a list in round robin distribution by the processors • Tests show that good load balancing is achieved by distributing units of work in a round robin fashion. • Benchmarks show almost linear scaling on thousands of files • Database file (~30 Mb) currently read in once per input file (one test case has ~30, 000 input file ; ~1000 procs) • Future plan : once per MPI process (reduces I/O from ~ 1 Tb to 30 Gb); eventually database file will be read once by single node SP Sci. Comp 6, Berkeley, August 23, 2002

Power 3 Timing and Speedup # files # proc Time (sec) Speedup 2227 32 2724 – 3031 1. 0 – 1. 0 2227 64 5531 – 5744 2. 0 – 1. 89 SP Sci. Comp 6, Berkeley, August 23, 2002

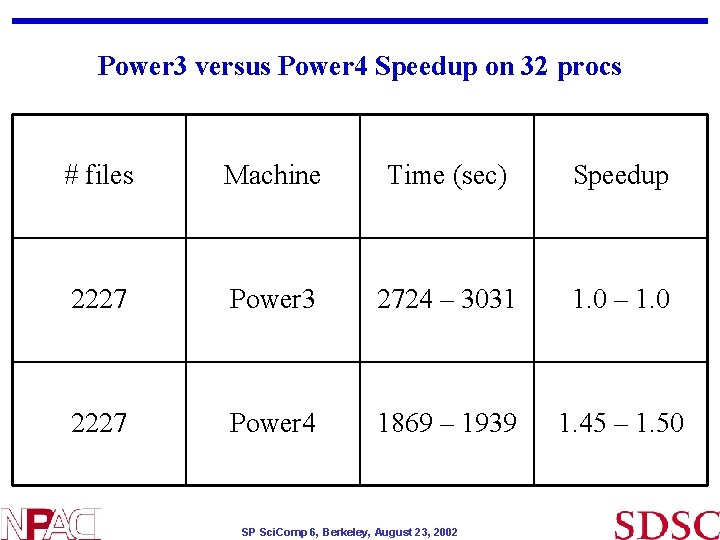
Power 3 versus Power 4 Speedup on 32 procs # files Machine Time (sec) Speedup 2227 Power 3 2724 – 3031 1. 0 – 1. 0 2227 Power 4 1869 – 1939 1. 45 – 1. 50 SP Sci. Comp 6, Berkeley, August 23, 2002

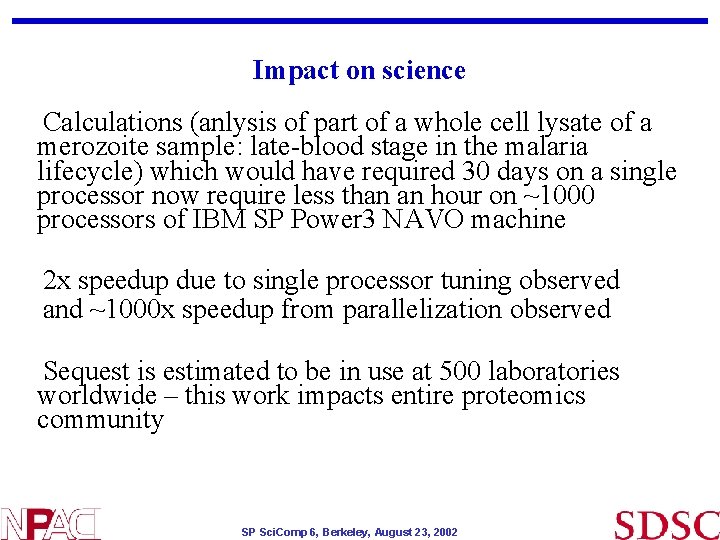
Impact on science Calculations (anlysis of part of a whole cell lysate of a merozoite sample: late-blood stage in the malaria lifecycle) which would have required 30 days on a single processor now require less than an hour on ~1000 processors of IBM SP Power 3 NAVO machine 2 x speedup due to single processor tuning observed and ~1000 x speedup from parallelization observed Sequest is estimated to be in use at 500 laboratories worldwide – this work impacts entire proteomics community SP Sci. Comp 6, Berkeley, August 23, 2002

Final Comments • Genomics community has already extensively used supercomputing capabilities and will continue to use • Now the proteomics community will use supercomputing more and more • Medical community is starting to use parallel computing in clinical and operation room procedures such as imaging, modeling of physical organs and their functions etc. SP Sci. Comp 6, Berkeley, August 23, 2002