Parallel Computation in Biological Sequence Analysis Par Align
Parallel Computation in Biological Sequence Analysis: Par. Align & Turbo. BLAST Larissa Smelkov
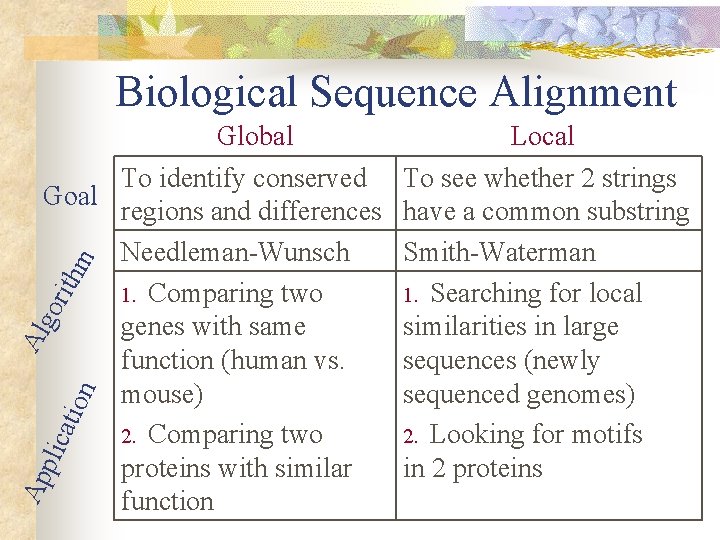
Biological Sequence Alignment Ap plic atio n Al go rith m Global To identify conserved Goal regions and differences Needleman-Wunsch 1. Comparing two genes with same function (human vs. mouse) 2. Comparing two proteins with similar function Local To see whether 2 strings have a common substring Smith-Waterman 1. Searching for local similarities in large sequences (newly sequenced genomes) 2. Looking for motifs in 2 proteins

Protein Responsible for Iron Transport Human MQEYTNHSDTTFALRNISFRVPGRTLLHPLSLTFPAGKVTGLIGHNGSGKSTLLK MLGRHPPSEGEILLDAQPLESWSSKAFARKVAYLPQQLPPAEGMTVRELVAIGR YPWHGALGRFGAADREKVEEAISLVGLKPLAHRLVDSLSGGERQRAWIAMLVA QDSRCLLLDEPTSALDIHQVDVLSLVHRLSQERGLTVIAVLHDINMAARYCDYL VALRGGEMIAQGTPAEIMRGETLEMIYGIPMGILPHPAGAAPVSFVY Chicken MKLILCTVLSLGIAAVCFAAPPKSVIRWCTISSPEEKKCNNLRDLTQQERISLTCV QKATYLDCIKAIANNEADAISLDGGQVFEAGLAPYKLKPIAAEIYEHTEGSTTSY YAVAVVKKGTEFTVNDLQGKTSCHTGLGRSAGWNIPIGTLLHWGAIEWEGIES GSVEQAVAKFFSASCVPGATIEQKLCRQCKGDPKTKCARNAPYSGYSGAFHCL KDGKGDVAFVKHTTVNENAPDLNDEYELLCLDGSRQPVDNYKTCNWARVAA HAVVARDDNKVEDIWSFLSKAQSDFGVDTKSDFHLFGPPGKKDPVLKDLLFKD SAIMLKRVPSLMSQLYLGFEYYSAIQSMRKDQLSGSPRQNRIQWIAVLKAEKSK CDRWSVVSNGDVECTVVDETKDCIIKIMKGEADAV

Protein Responsible for Iron Transport Human MQEYTNHSDTTFALRNISFRVPGRTLLHPLSLTFPAGKVTGLIGHNGSGKSTLLK MLGRHPPSEGEILLDAQPLESWSSKAFARKVAYLPQQLPPAEGMTVRELVAIGR YPWHGALGRFGAADREKVEEAISLVGLKPLAHRLVDSLSGGERQRAWIAMLVA QDSRCLLLDEPTSALDIHQVDVLSLVHRLSQERGLTVIAVLHDINMAARYCDYL VALRGGEMIAQGTPAEIMRGETLEMIYGIPMGILPHPAGAAPVSFVY Chicken MKLILCTVLSLGIAAVCFAAPPKSVIRWCTISSPEEKKCNNLRDLTQQERISLTCV QKATYLDCIKAIANNEADAISLDGGQVFEAGLAPYKLKPIAAEIYEHTEGSTTSY YAVAVVKKGTEFTVNDLQGKTSCHTGLGRSAGWNIPIGTLLHWGAIEWEGIES GSVEQAVAKFFSASCVPGATIEQKLCRQCKGDPKTKCARNAPYSGYSGAFHCL KDGKGDVAFVKHTTVNENAPDLNDEYELLCLDGSRQPVDNYKTCNWARVAA HAVVARDDNKVEDIWSFLSKAQSDFGVDTKSDFHLFGPPGKKDPVLKDLLFKD SAIMLKRVPSLMSQLYLGFEYYSAIQSMRKDQLSGSPRQNRIQWIAVLKAEKSK CDRWSVVSNGDVECTVVDETKDCIIKIMKGEADAV

Similar Substrings DSLSGGERQ–RA–WIAMLVAQDSRC : : : : DQLSGSPRQNRIQWIAVLKAEKSKC

Talk Outline n n n Problem Description Smith-Waterman Algorithm BLAST Par. Align Turbo. BLAST Comparison

Problems of Comparison of 2 Sequences n Evolution Factor n n Additions Deletions Substitutions Human Factor n n Typos Duplicates

Solution n Smith-Waterman Algorithm (S-W) Score Matrix Gap Penalty
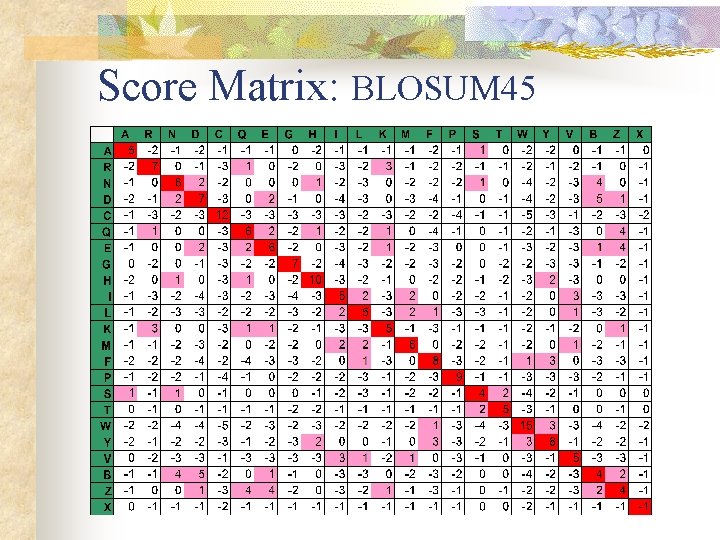
Score Matrix: BLOSUM 45

Pairwise Alignment Example ELEPHANT PANTHER

S-W: Dynamic Programming Matrix
![S-W: Formula T[i, j] = max T[i-1, j-1] + score(s[i], t[j]) T[i-1, j] – S-W: Formula T[i, j] = max T[i-1, j-1] + score(s[i], t[j]) T[i-1, j] –](http://slidetodoc.com/presentation_image_h2/346c8d704e2e7507edd69ae27dc3a25f/image-12.jpg)
S-W: Formula T[i, j] = max T[i-1, j-1] + score(s[i], t[j]) T[i-1, j] – g T[i, j-1] – g 0 g – gap penalty g = 8 (in our example) T[i-1, j-1] T[i-1, j] ?

S-W: Dynamic Programming Matrix

S-W: Dynamic Programming Matrix

S-W: Dynamic Programming Matrix

S-W: Dynamic Programming Matrix

S-W: Result Alignment ELEPHANT : : P– ANTHER

S-W: Summary n Uses n n n Complexity n n Score matrix Gap penalties O(mn) Sensitivity n High
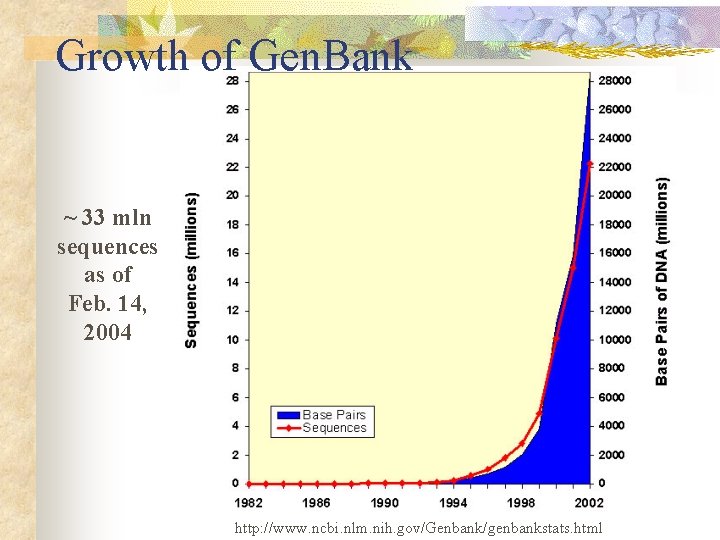
Growth of Gen. Bank ~ 33 mln sequences as of Feb. 14, 2004 http: //www. ncbi. nlm. nih. gov/Genbank/genbankstats. html

BLAST: Basic Local Alignment Search Tool

BLAST: Steps n Divide both sequences into words of length w n n n default w = 3 Calculate score for each pair Extend high scored pairs to increase score

BLAST: Divide Sequences
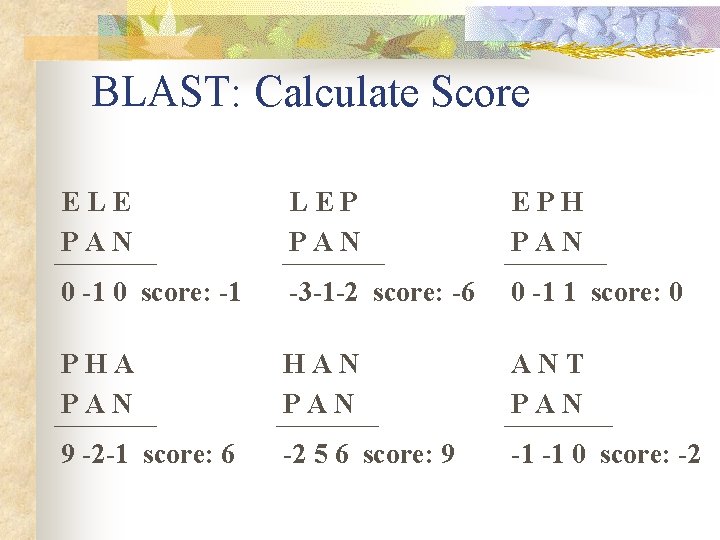
BLAST: Calculate Score ELE PAN LEP PAN EPH PAN 0 -1 0 score: -1 -3 -1 -2 score: -6 0 -1 1 score: 0 PHA PAN HAN PAN ANT PAN 9 -2 -1 score: 6 -2 5 6 score: 9 -1 -1 0 score: -2
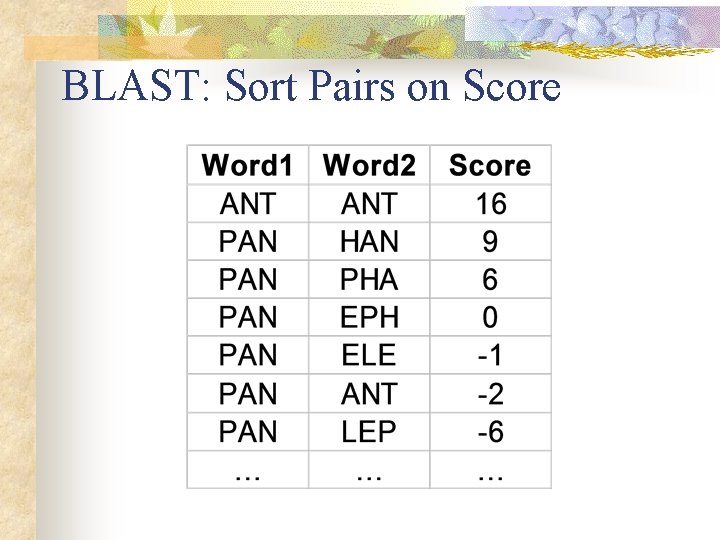
BLAST: Sort Pairs on Score
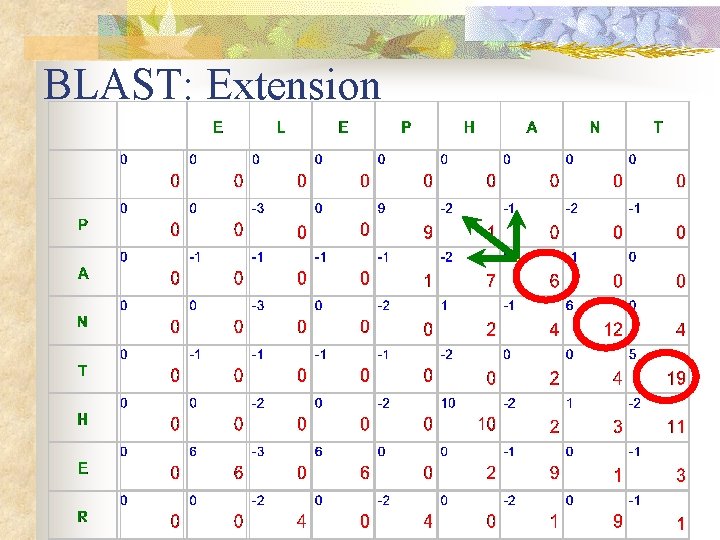
BLAST: Extension

BLAST: Summary n Uses n n Complexity n n Score matrix Gap penalties Heuristics to reduce computations O(m) with O(n) processors Sensitivity n Low

Sensitivity n n n Task: Align 2 sequences: AXBXCXDXE ABCDE Smith-Waterman: AXBXCXDXE : : : A– B– C– D– E BLAST: Ø (no similar substrings)
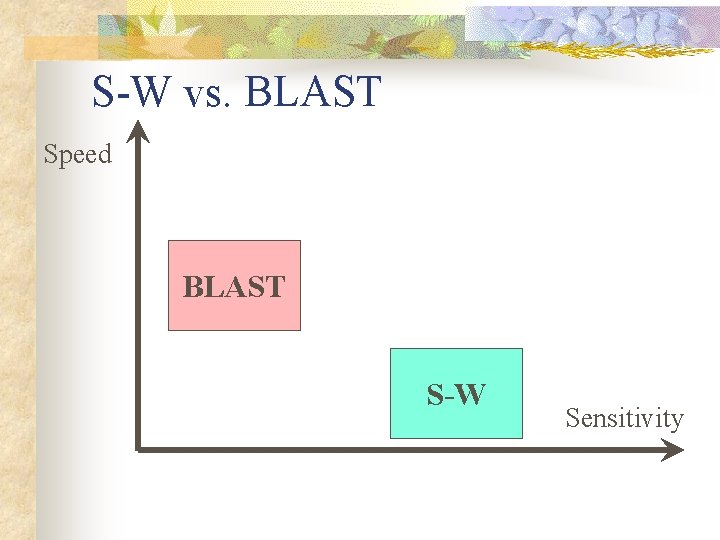
S-W vs. BLAST Speed BLAST S-W Sensitivity

S-W and BLAST n Using them now n n Too costly Inefficient Time-consuming Solution n n More heuristics More parallelism

Par. Align

Par. Align: Steps n n Find ungapped alignments Calculate approximate alignment scores Choose high-scored sequences Apply S-W

Par. Align: Microparallelism n n n Divide wide registers into smaller units Perform the same operation on different data sources Modern microprocessors have this technology built in

Par. Align: Calculate Scores in Parallel
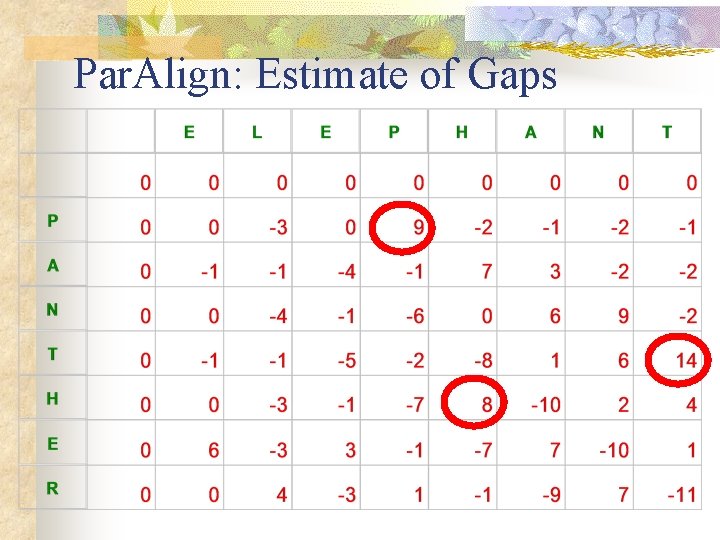
Par. Align: Estimate of Gaps
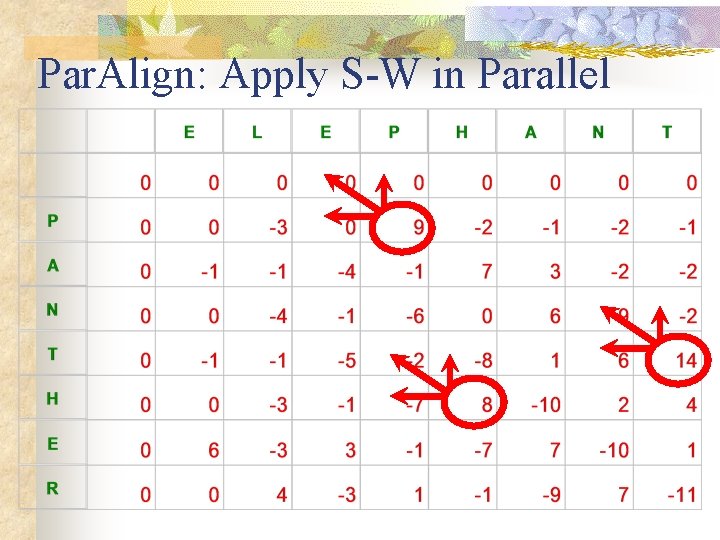
Par. Align: Apply S-W in Parallel

Par. Align: Summary n Uses n n Requirement for machine n n Modern microprocessor Speed n n SIMD technology (single instruction multiple data) S-W Algorithm Heuristics to reduce computations Fast Sensitivity n Medium

Turbo. BLAST

Turbo. BLAST: Steps n Divide the job n n n Parts of query against partition of database Apply BLAST Merge results
Turbo. BLAST: Implementation n n A three-tier system Components n n n Client Master Workers
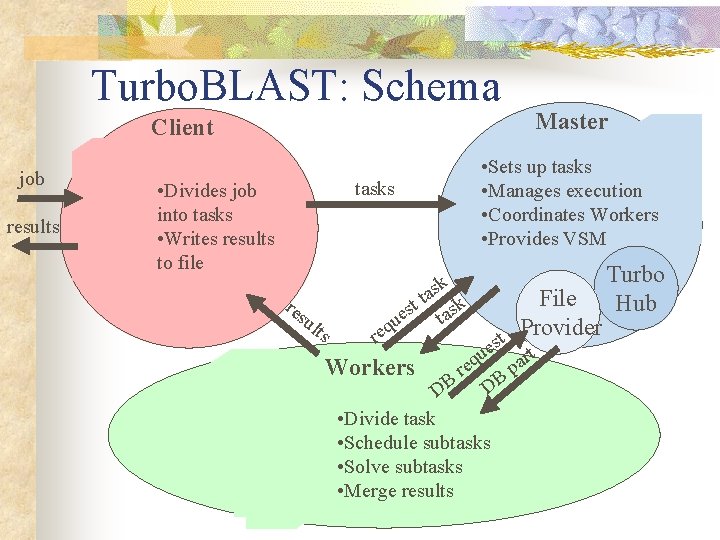
Turbo. BLAST: Schema Client job results • Sets up tasks • Manages execution • Coordinates Workers • Provides VSM tasks • Divides job into tasks • Writes results to file res ult s k s a t t sk s ta ue req Workers Master File Provider t es rt u pa req B DB D • Divide task • Schedule subtasks • Solve subtasks • Merge results Turbo Hub

Turbo. BLAST: Client n n n Takes a BLAST job and divides it into a number of initial BLAST tasks. Submits these tasks to the Master Retrieves the results, and writes them to file.
Turbo. BLAST: Master n n n Accepts tasks from Clients and sets them up to for processing by the Workers Includes Turbo. Hub (the server portion of a parallel execution system) Includes File Provider (Java application that manages the databases)

Turbo. BLAST: Worker n n Workers are processors Run a Java application and perform the BLAST computations Merge the result Are responsible for scheduling
Turbo. Hub n n n n Turbo. Hub is execution engine for parallel and distributed Java applications Scalable high performance Wide range of computing environments Manages the flow of data through the workflows Schedules the components Transforms data between components Balances load Handles errors

Turbo. BLAST: Turbo. Hub n n n Manages task execution Coordinates the Workers Provides a virtual shared memories Supports dynamic changes in the set of Workers Supports fault tolerance

Turbo. BLAST: File Provider n n Maintains a copy of each database Delivers all or part of each database to Workers as they require them

Turbo. BLAST: Advantages n Size of each task is optimal n n Large set of tasks n n no waste of time for processors No algorithm change n n n processing is efficient on the processor that computes the task Support for all flavors of BLAST Ease to update Applicable for different environments (PC, Macintosh …)

Turbo. BLAST: Experiment n Input data n n n Database n n 500 proteins 200 – 400 amino acids in each 1, 681, 522, 266 sequences Hardware n n IBM Linux cluster 8 dual-processor workstations n n n 2 Pentium III processors, 996 Mhz each 2 Gbyte memory 100 Mbit Ethernet
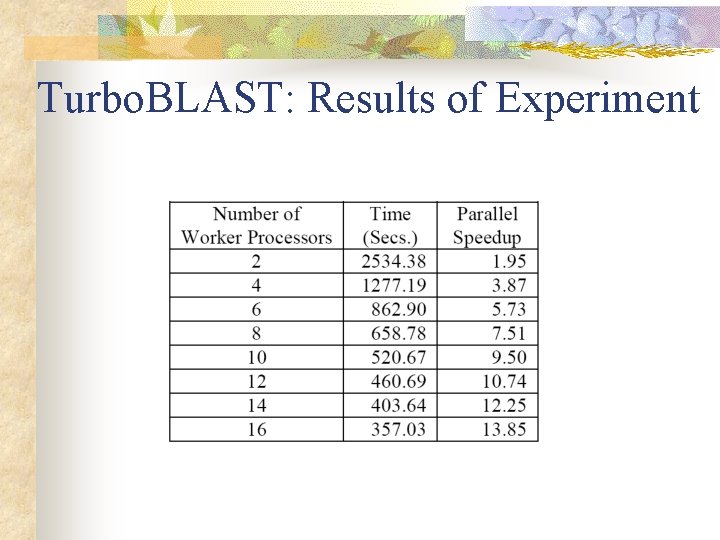
Turbo. BLAST: Results of Experiment
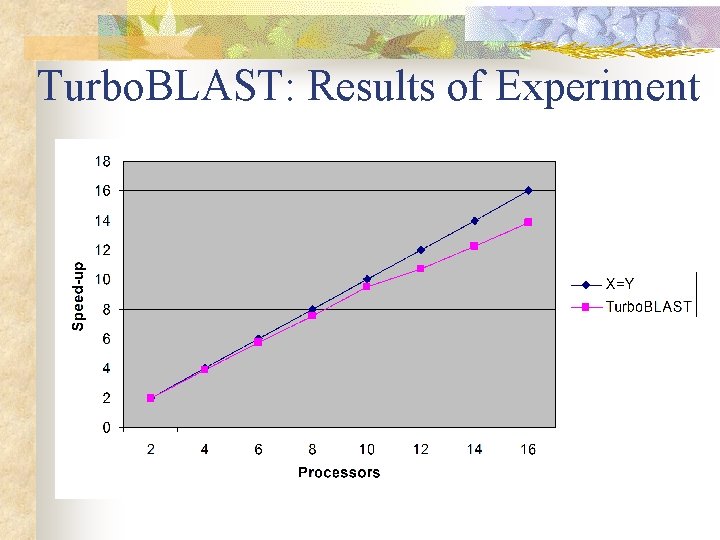
Turbo. BLAST: Results of Experiment

Turbo. BLAST: Summary n Divide and Conquer n n n Uses BLAST Algorithm Requirement for each machine n n n Java VM Local BLAST executable Speed n n Use many copies of BLAST in parallel Very fast Sensitivity n Low
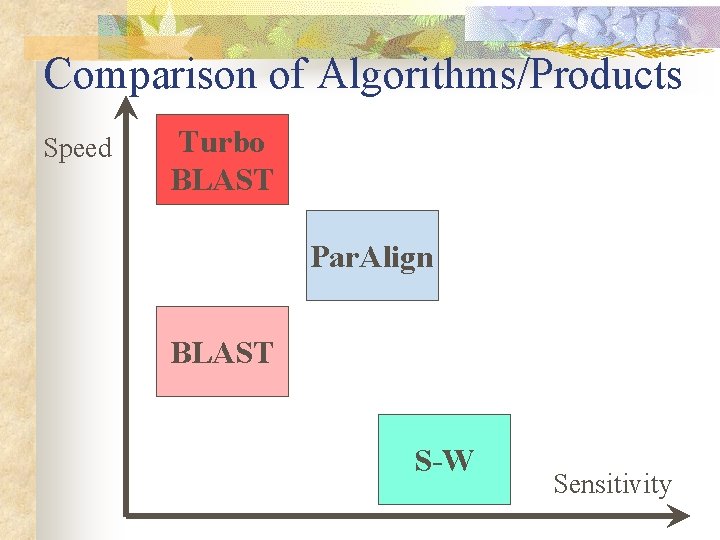
Comparison of Algorithms/Products Speed Turbo BLAST Par. Align BLAST S-W Sensitivity
References n n R. D. Bjornson, A. H. Sherman, S. B. Weston, N. Willard, J. Wing “Turbo. BLAST: A Parallel Implementation of BLAST Built on the Turbo. Hub” Intl. Parallel and Distributed Processing Symposium (IPDPS), 2002. Rognes T. “Par. Align: a parallel sequence alignment algorithm for rapid and sensitive database searches” Oxford University Press, 2001

Don’t ask any Questions, please…

PS n Web site there you can donate your computer time to participate in search of methods to cure cancer: http: //www. the-optimists. org. uk
- Slides: 55