Pairwise alignment n n n Now we know



- Slides: 22

Pairwise alignment n n n Now we know how to do it: How do we get a multiple alignment (three or more sequences)? Multiple alignment: much greater combinatorial explosion than with pairwise alignment…. .

Multi-dimensional dynamic programming (Murata et al. 1985)

Simultaneous Multiple alignment Multi-dimensional dynamic programming MSA (Lipman et al. , 1989, PNAS 86, 4412) n extremely slow and memory intensive n up to 8 -9 sequences of ~250 residues DCA (Stoye et al. , 1997, CABIOS 13, 625) n still very slow

Alternative multiple alignment methods Biopat (first method ever) u MULTAL (Taylor 1987) u DIALIGN (Morgenstern 1996) u PRRP (Gotoh 1996) u Clustal (Thompson Higgins Gibson 1994) u Praline (Heringa 1999) u T Coffee (Notredame 2000) u HMMER (Eddy 1998) [Hidden Marcov Models] u SAGA (Notredame 1996) [Genetic algorithms] u

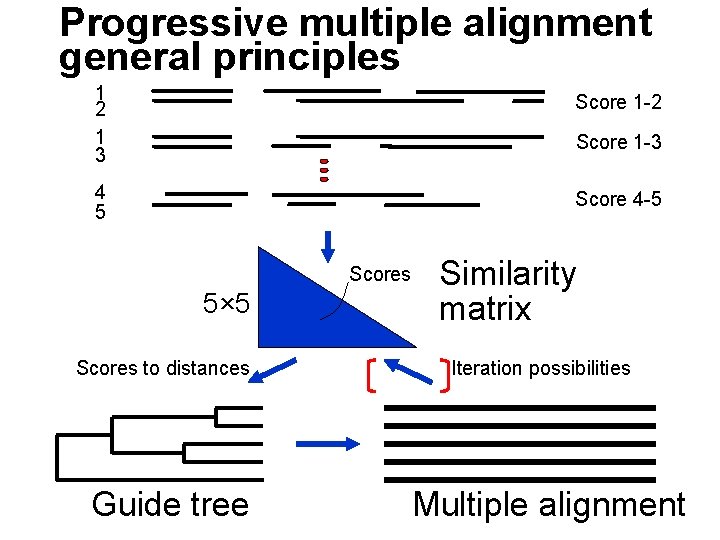
Progressive multiple alignment general principles 1 2 1 3 Score 1 -2 4 5 Score 4 -5 Score 1 -3 Scores 5× 5 Scores to distances Guide tree Similarity matrix Iteration possibilities Multiple alignment

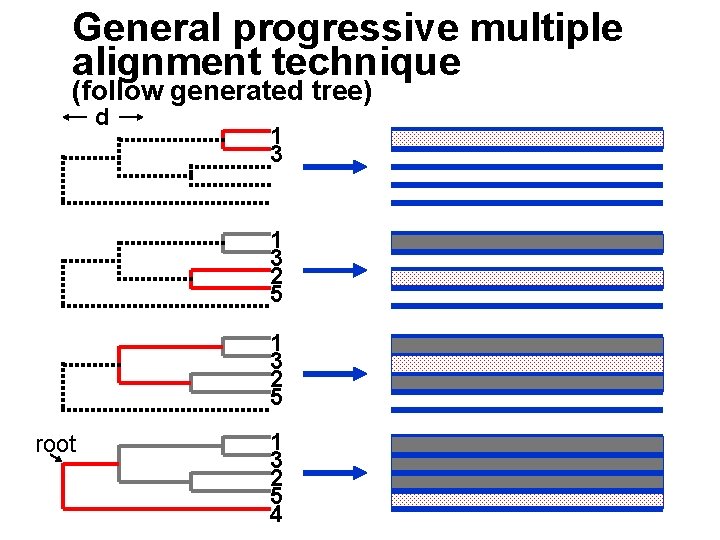
General progressive multiple alignment technique (follow generated tree) d 1 3 2 5 root 1 3 2 5 4

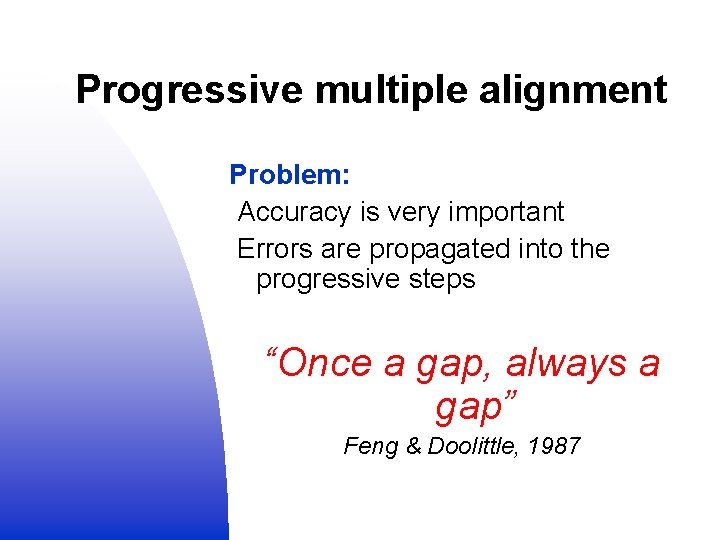
Progressive multiple alignment Problem: Accuracy is very important Errors are propagated into the progressive steps “Once a gap, always a gap” Feng & Doolittle, 1987

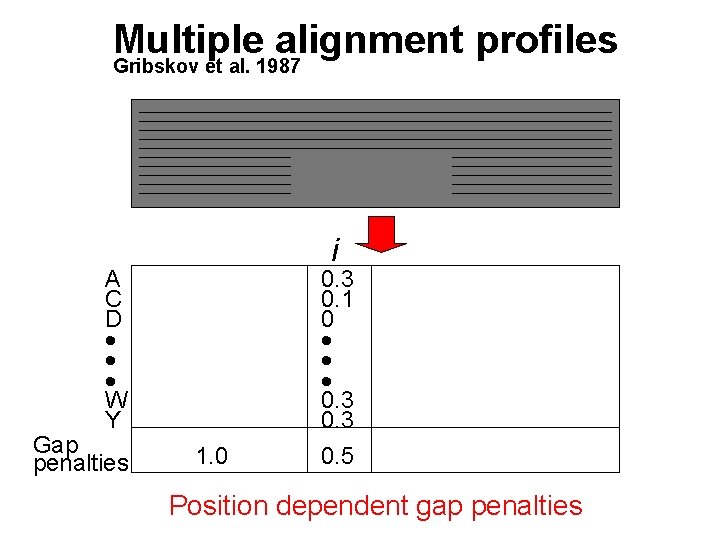
Multiple alignment profiles Gribskov et al. 1987 i A C D W Y Gap penalties 1. 0 0. 3 0. 1 0 0. 3 0. 5 Position dependent gap penalties

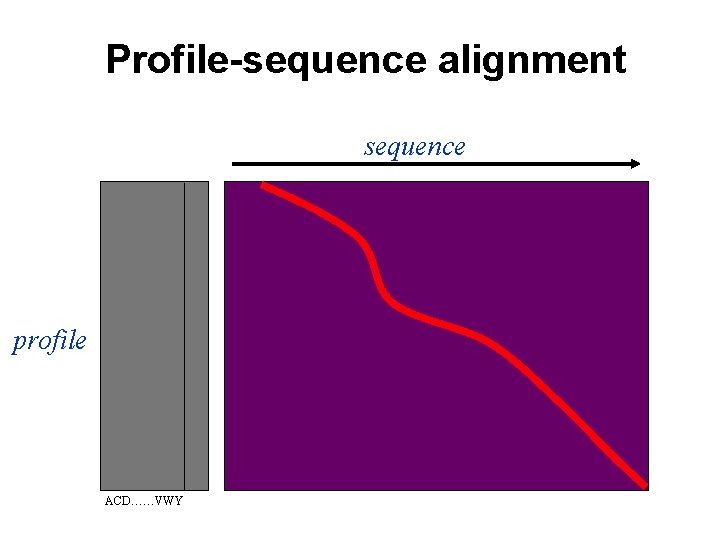
Profile-sequence alignment sequence profile ACD……VWY

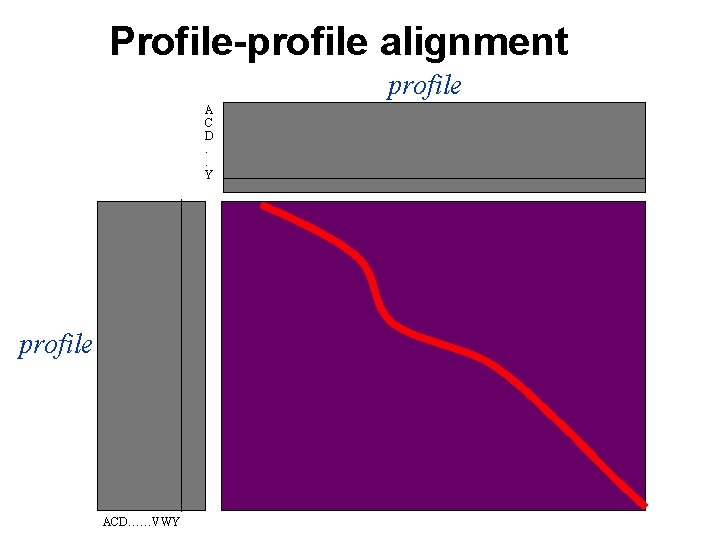
Profile-profile alignment profile A C D. . Y profile ACD……VWY

Clustal, Clustal. W, Clustal. X n n n CLUSTAL W/X (Thompson et al. , 1994) uses Neighbour Joining (NJ) algorithm (Saitou and Nei, 1984), widely used in phylogenetic analysis, to construct guide tree. Sequence blocks are represented by profiles, in which the individual sequences are additionally weighted according to the branch lengths in the NJ tree. Further carefully crafted heuristics include: u u u n (i) local gap penalties (ii) automatic selection of the amino acid substitution matrix, (iii) automatic gap penalty adjustment (iv) mechanism to delay alignment of sequences that appear to be distant at the time they are considered. CLUSTAL (W/X) does not allow iteration (Hogeweg and Hesper, 1984; Corpet, 1988, Gotoh, 1996; Heringa, 1999, 2002)

Strategies for multiple sequence alignment n n Profile pre-processing Secondary structure-induced alignment Globalised local alignment Matrix extension Objective: try to avoid (early) errors

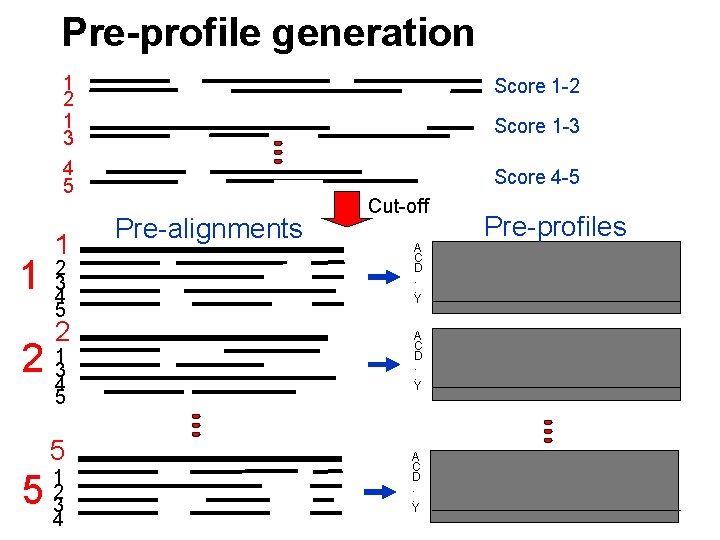
Pre-profile generation 1 1 2 1 3 Score 1 -2 4 5 Score 4 -5 1 2 3 4 5 2 2 134 5 5 5 1 2 3 4 Score 1 -3 Pre-alignments Cut-off A C D. . Y Pre-profiles

Strategies for multiple sequence alignment n n Profile pre-processing Secondary structure-induced alignment Globalised local alignment Matrix extension Objective: try to avoid (early) errors

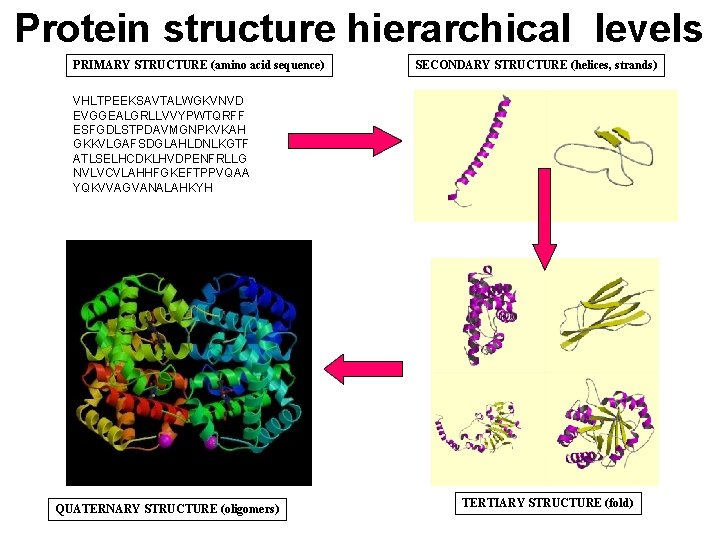
Protein structure hierarchical levels PRIMARY STRUCTURE (amino acid sequence) SECONDARY STRUCTURE (helices, strands) VHLTPEEKSAVTALWGKVNVD EVGGEALGRLLVVYPWTQRFF ESFGDLSTPDAVMGNPKVKAH GKKVLGAFSDGLAHLDNLKGTF ATLSELHCDKLHVDPENFRLLG NVLVCVLAHHFGKEFTPPVQAA YQKVVAGVANALAHKYH QUATERNARY STRUCTURE (oligomers) TERTIARY STRUCTURE (fold)

Strategies for multiple sequence alignment n Profile pre-processing Secondary structure-induced alignment n Globalised local alignment n Matrix extension n Objective: try to avoid (early) errors

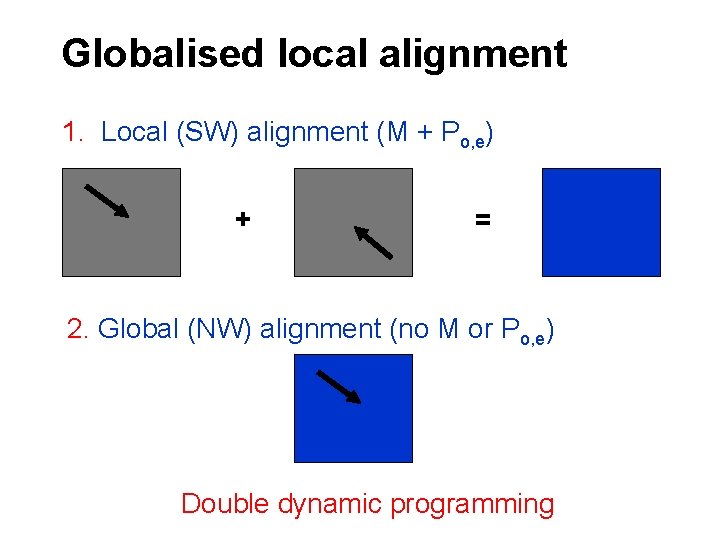
Globalised local alignment 1. Local (SW) alignment (M + Po, e) + = 2. Global (NW) alignment (no M or Po, e) Double dynamic programming

Strategies for multiple sequence alignment n Profile pre-processing Secondary structure-induced alignment Globalised local alignment n Matrix extension n n Objective: try to avoid (early) errors

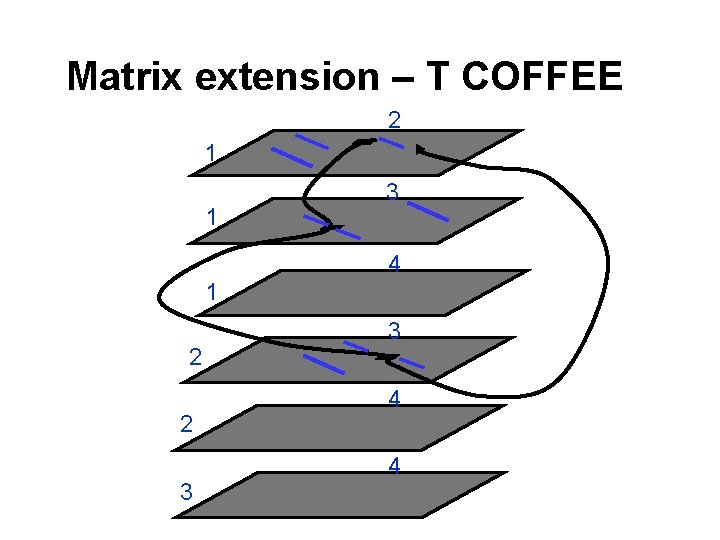
Matrix extension – T COFFEE 2 1 1 3 4 1 2 2 3 3 4 4

Summary n Weighting schemes simulating simultaneous multiple alignment Profile pre-processing (global/local) u Matrix extension (well balanced scheme) u n Smoothing alignment signals u n Using additional information u n globalised local alignment secondary structure driven alignment Schemes strike balance between speed and sensitivity

References n n n Heringa, J. (1999) Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comp. Chem. 23, 341 -364. Notredame, C. , Higgins, D. G. , Heringa, J. (2000) TCoffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. , 302, 205 -217. Heringa, J. (2002) Local weighting schemes for protein multiple sequence alignment. Comput. Chem. , 26(5), 459 -477.

Where to find this…. http: //www. cs. vu. nl/~ibivu/teaching