Packing Cavities atomic radii contactdistance profiles cavities PP

- Slides: 23

Packing, Cavities • atomic radii, contactdistance profiles • cavities • P-P interactions • crystal contacts • solvent channels • de-stabilizing mutations in core (TS mutants? ) • entropy effects on surface

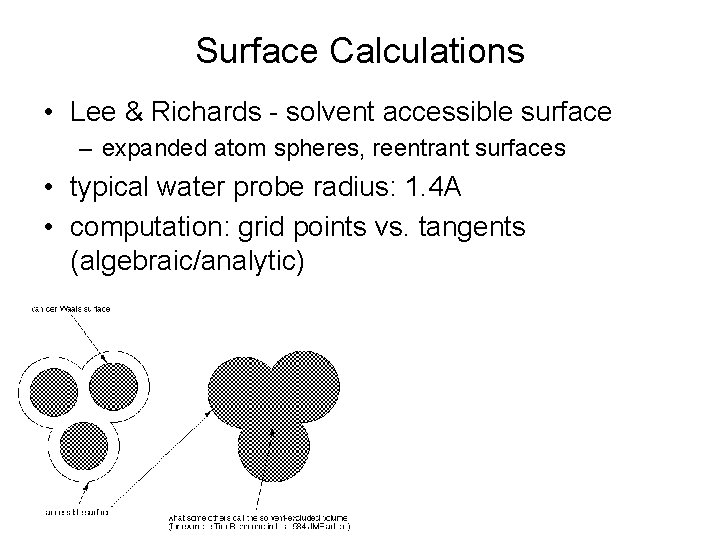
Surface Calculations • Lee & Richards - solvent accessible surface – expanded atom spheres, reentrant surfaces • typical water probe radius: 1. 4 A • computation: grid points vs. tangents (algebraic/analytic)

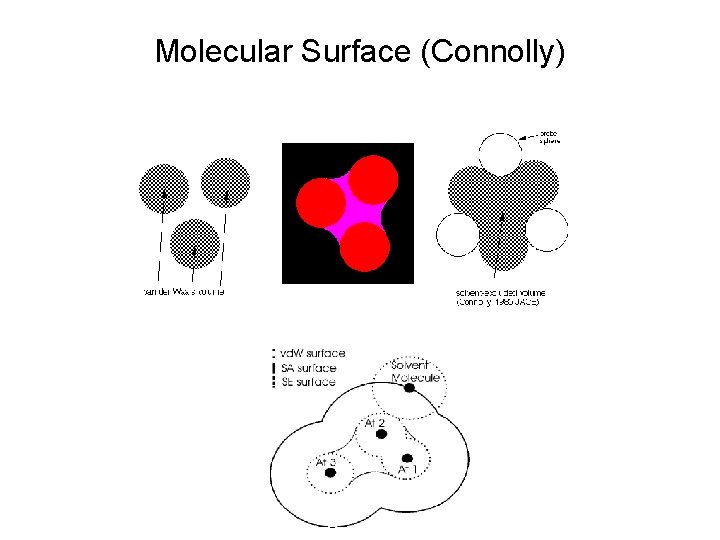
Molecular Surface (Connolly)

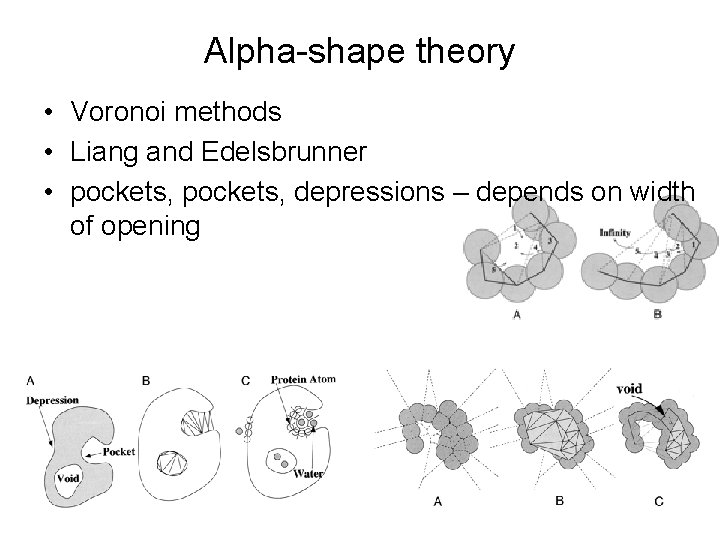
Alpha-shape theory • Voronoi methods • Liang and Edelsbrunner • pockets, depressions – depends on width of opening

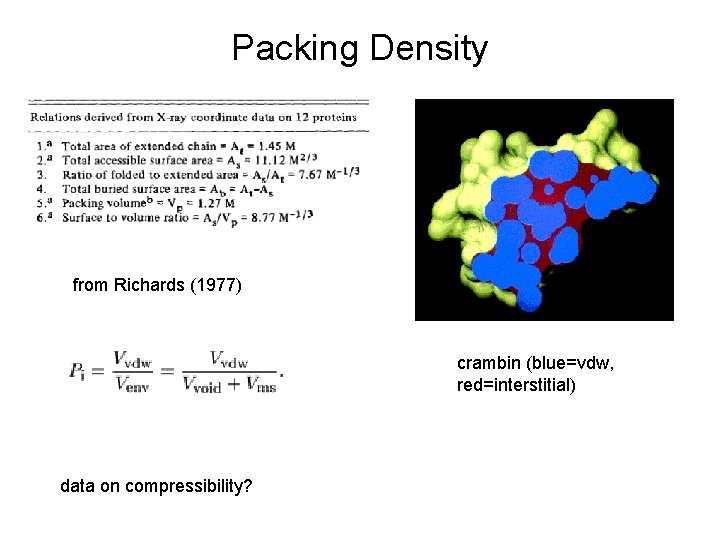
Packing Density from Richards (1977) crambin (blue=vdw, red=interstitial) data on compressibility?

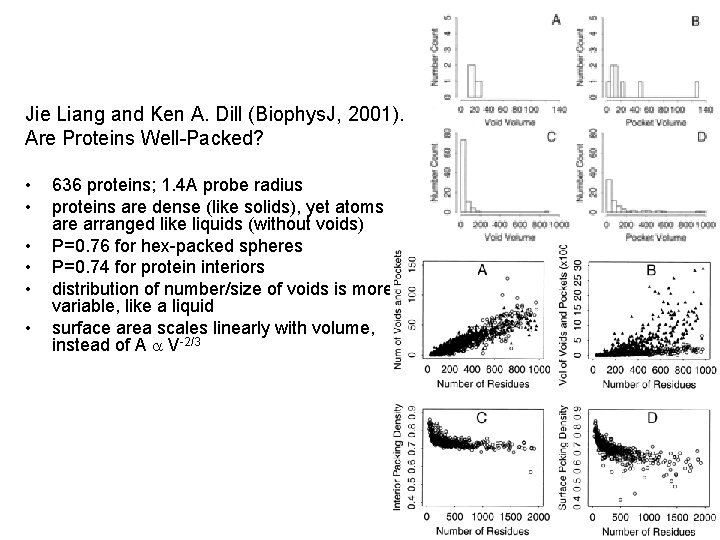
Jie Liang and Ken A. Dill (Biophys. J, 2001). Are Proteins Well-Packed? • • • 636 proteins; 1. 4 A probe radius proteins are dense (like solids), yet atoms are arranged like liquids (without voids) P=0. 76 for hex-packed spheres P=0. 74 for protein interiors distribution of number/size of voids is more variable, like a liquid surface area scales linearly with volume, instead of A a V-2/3

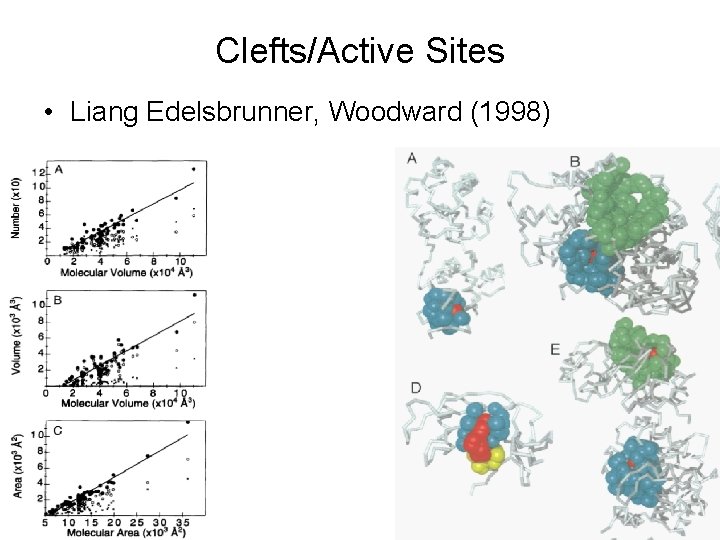
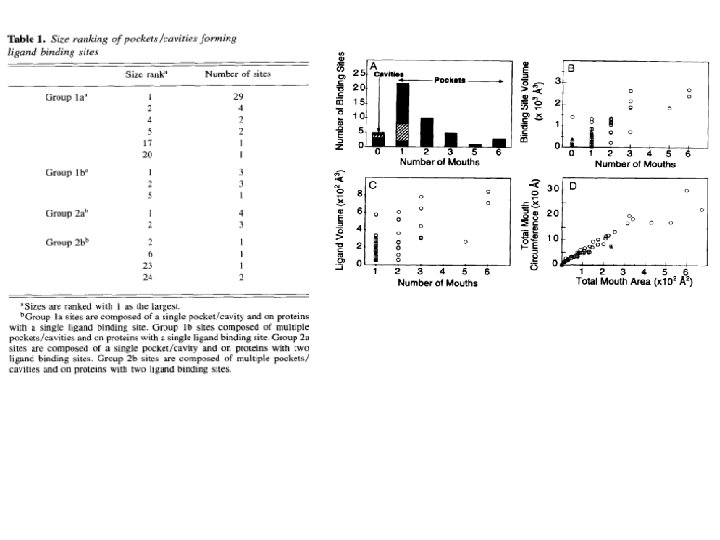
Clefts/Active Sites • Liang Edelsbrunner, Woodward (1998)


• Laskowski, Luscombe, Swindells, and Thornton (1996)

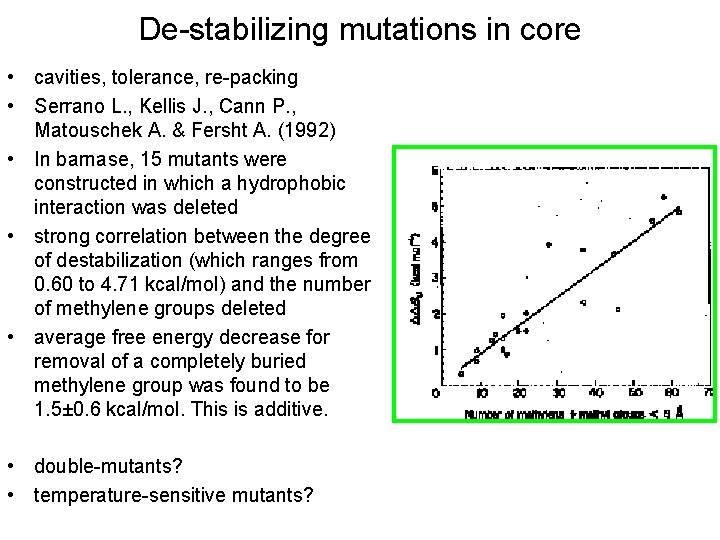
De-stabilizing mutations in core • cavities, tolerance, re-packing • Serrano L. , Kellis J. , Cann P. , Matouschek A. & Fersht A. (1992) • In barnase, 15 mutants were constructed in which a hydrophobic interaction was deleted • strong correlation between the degree of destabilization (which ranges from 0. 60 to 4. 71 kcal/mol) and the number of methylene groups deleted • average free energy decrease for removal of a completely buried methylene group was found to be 1. 5± 0. 6 kcal/mol. This is additive. • double-mutants? • temperature-sensitive mutants?

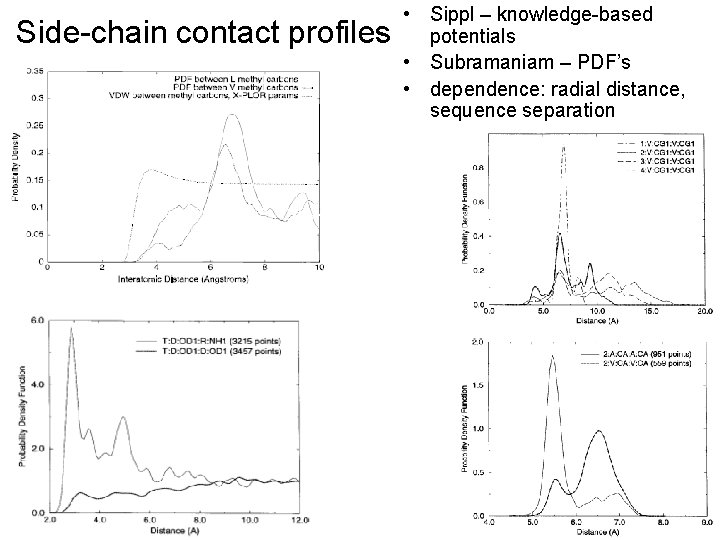
Side-chain contact profiles • Sippl – knowledge-based potentials • Subramaniam – PDF’s • dependence: radial distance, sequence separation

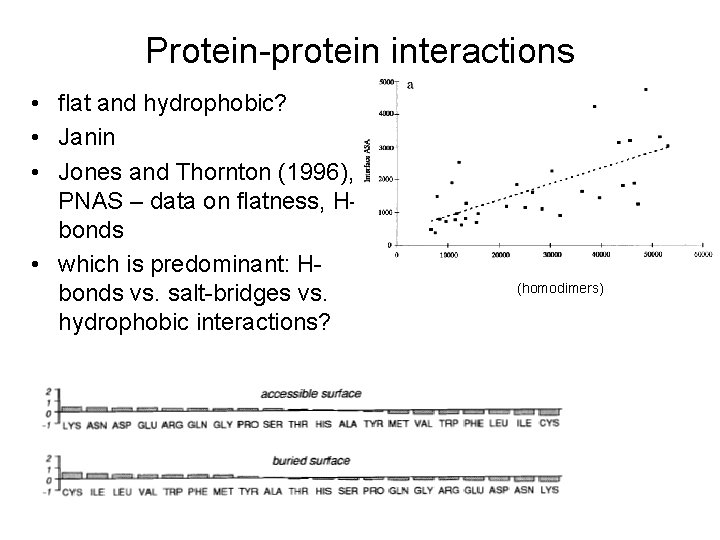
Protein-protein interactions • flat and hydrophobic? • Janin • Jones and Thornton (1996), PNAS – data on flatness, Hbonds • which is predominant: Hbonds vs. salt-bridges vs. hydrophobic interactions? (homodimers)

Complementarity of P-P interfaces • shape complementarity – measure “gaps” or voids – cavities at interfaces (Hubbard and Argos, 1994) • more common than in core • suggests complementarity doesn’t have to be perfect – surface normals: • R Norel, SL Lin, HL Wolfson and R Nussinov

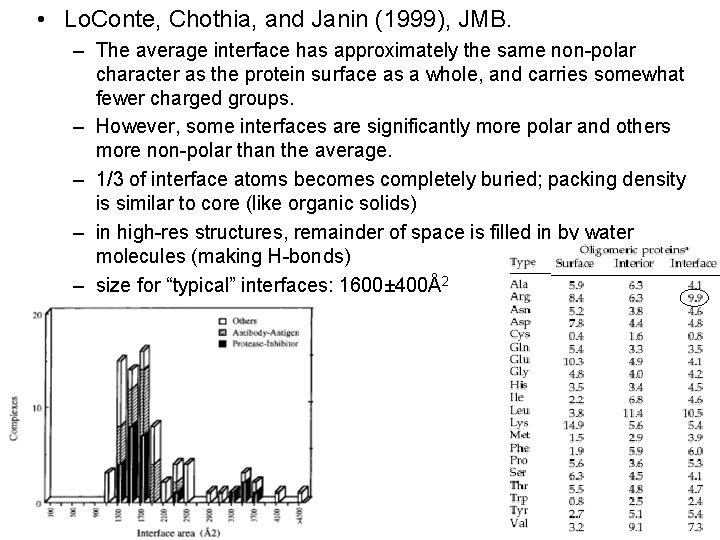
• Lo. Conte, Chothia, and Janin (1999), JMB. – The average interface has approximately the same non-polar character as the protein surface as a whole, and carries somewhat fewer charged groups. – However, some interfaces are significantly more polar and others more non-polar than the average. – 1/3 of interface atoms becomes completely buried; packing density is similar to core (like organic solids) – in high-res structures, remainder of space is filled in by water molecules (making H-bonds) – size for “typical” interfaces: 1600± 400Å2

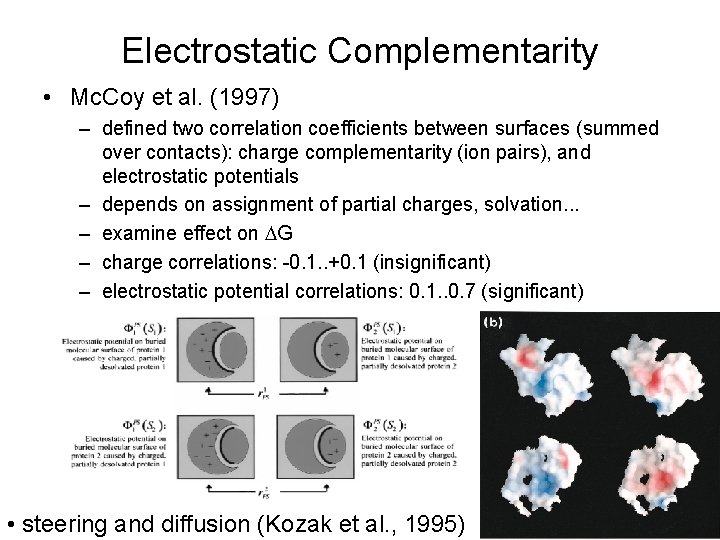
Electrostatic Complementarity • Mc. Coy et al. (1997) – defined two correlation coefficients between surfaces (summed over contacts): charge complementarity (ion pairs), and electrostatic potentials – depends on assignment of partial charges, solvation. . . – examine effect on DG – charge correlations: -0. 1. . +0. 1 (insignificant) – electrostatic potential correlations: 0. 1. . 0. 7 (significant) • steering and diffusion (Kozak et al. , 1995)

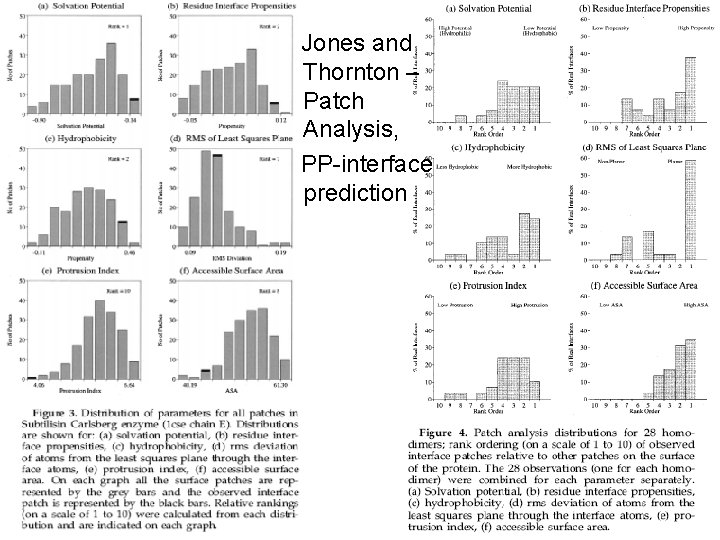
Jones and Thornton – Patch Analysis, PP-interface prediction

Examples of P-P interactions • • • b-lactamase/BLIP – one of the tightest antibody-antigens (HYHel 5) SH 2/SH 3 and tyrosine kinases PDZ domains calmodulin proteases, kinases (recognize+catalyze)

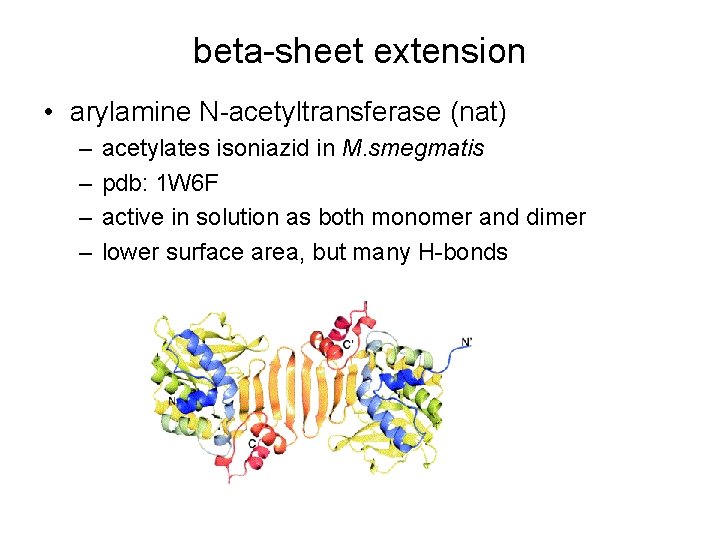
beta-sheet extension • arylamine N-acetyltransferase (nat) – – acetylates isoniazid in M. smegmatis pdb: 1 W 6 F active in solution as both monomer and dimer lower surface area, but many H-bonds

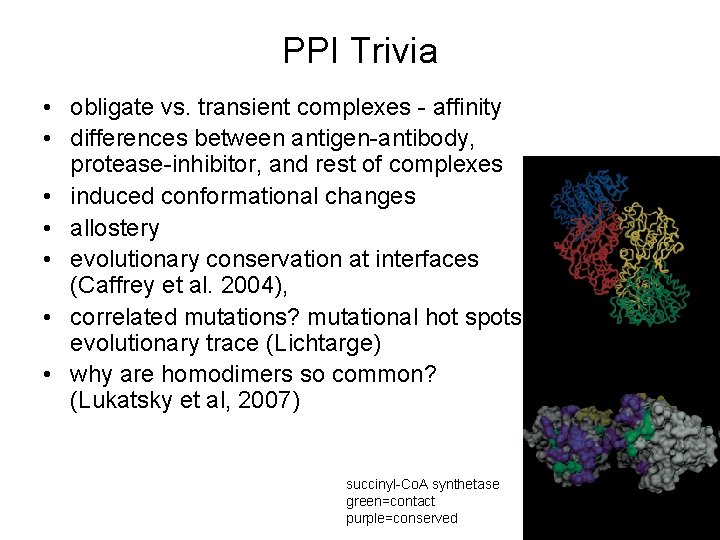
PPI Trivia • obligate vs. transient complexes - affinity • differences between antigen-antibody, protease-inhibitor, and rest of complexes • induced conformational changes • allostery • evolutionary conservation at interfaces (Caffrey et al. 2004), • correlated mutations? mutational hot spots, evolutionary trace (Lichtarge) • why are homodimers so common? (Lukatsky et al, 2007) succinyl-Co. A synthetase green=contact purple=conserved

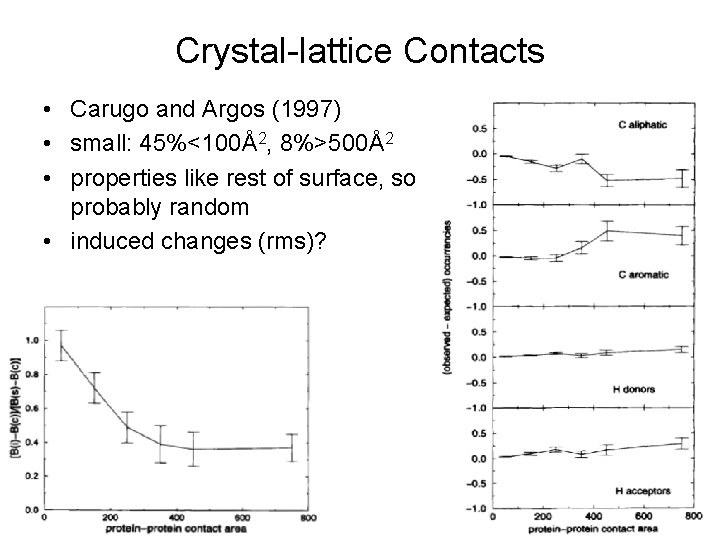
Crystal-lattice Contacts • Carugo and Argos (1997) • small: 45%<100Å2, 8%>500Å2 • properties like rest of surface, so probably random • induced changes (rms)?

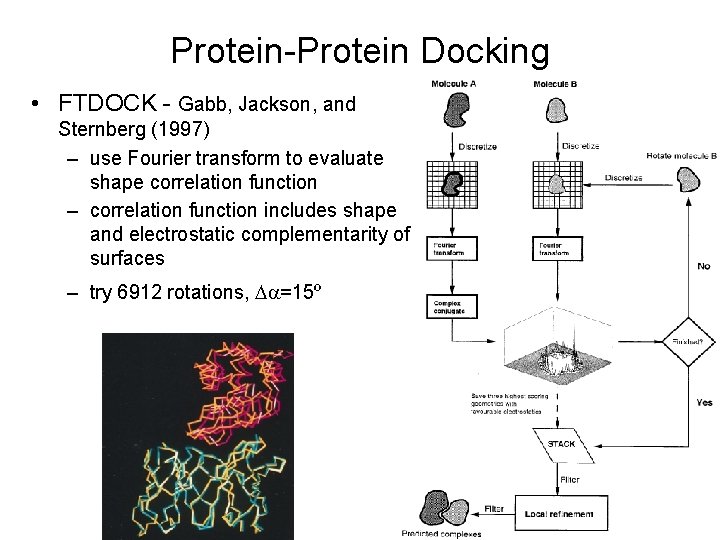
Protein-Protein Docking • FTDOCK - Gabb, Jackson, and Sternberg (1997) – use Fourier transform to evaluate shape correlation function – correlation function includes shape and electrostatic complementarity of surfaces – try 6912 rotations, Da=15º

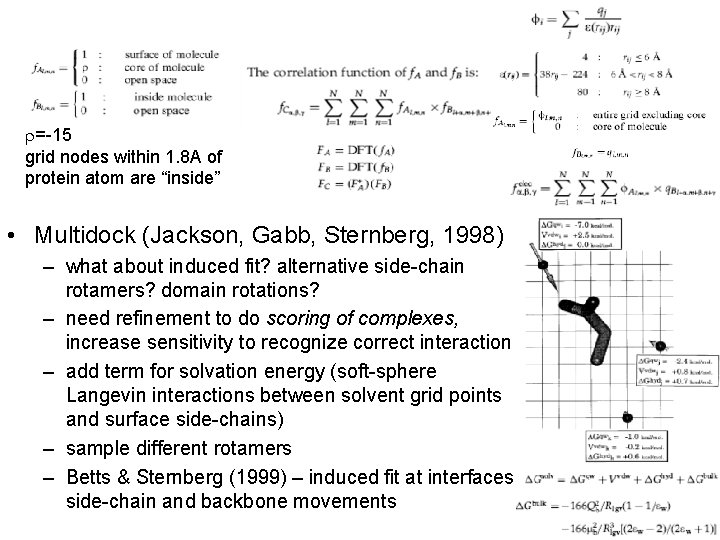
r=-15 grid nodes within 1. 8 A of protein atom are “inside” • Multidock (Jackson, Gabb, Sternberg, 1998) – what about induced fit? alternative side-chain rotamers? domain rotations? – need refinement to do scoring of complexes, increase sensitivity to recognize correct interaction – add term for solvation energy (soft-sphere Langevin interactions between solvent grid points and surface side-chains) – sample different rotamers – Betts & Sternberg (1999) – induced fit at interfaces side-chain and backbone movements

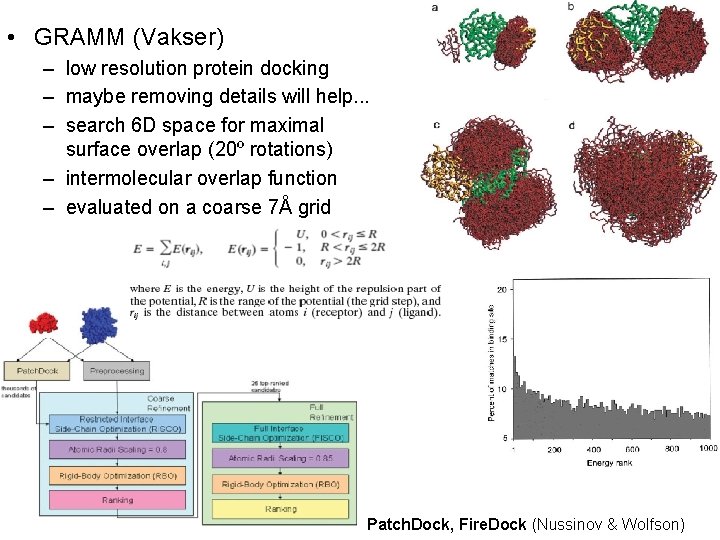
• GRAMM (Vakser) – low resolution protein docking – maybe removing details will help. . . – search 6 D space for maximal surface overlap (20º rotations) – intermolecular overlap function – evaluated on a coarse 7Å grid Patch. Dock, Fire. Dock (Nussinov & Wolfson)