Oxidative Phosphorylation Oxidative Phosphorylation Final step of cellular



![Experimental Support [K+] < [Cl-] Experimental Support [K+] < [Cl-]](https://slidetodoc.com/presentation_image_h2/9d0187dabf181bb00c67f00554c21b3e/image-36.jpg)

- Slides: 48

Oxidative Phosphorylation

Oxidative Phosphorylation Final step of cellular respiration n Convergence of pathways n Reduced cofactors oxidized n – Respiratory chain Oxidation coupled to phosphorylation n Occurs in mitochondria n Chemiosmotic theory n

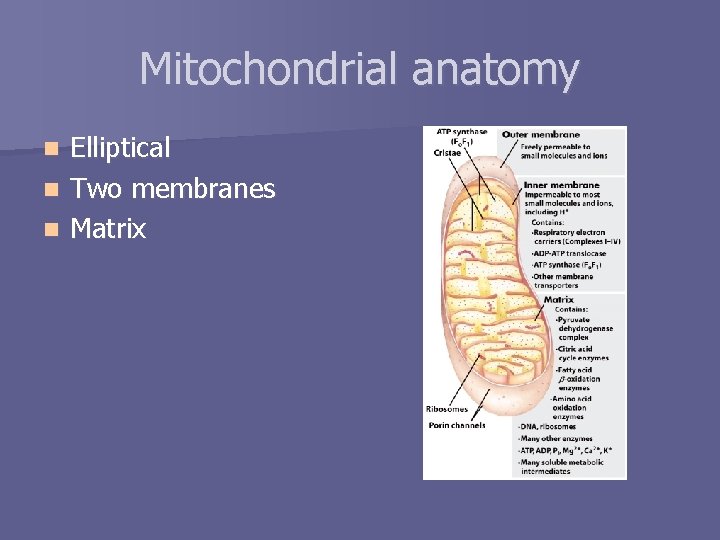
Mitochondrial anatomy Elliptical n Two membranes n Matrix n

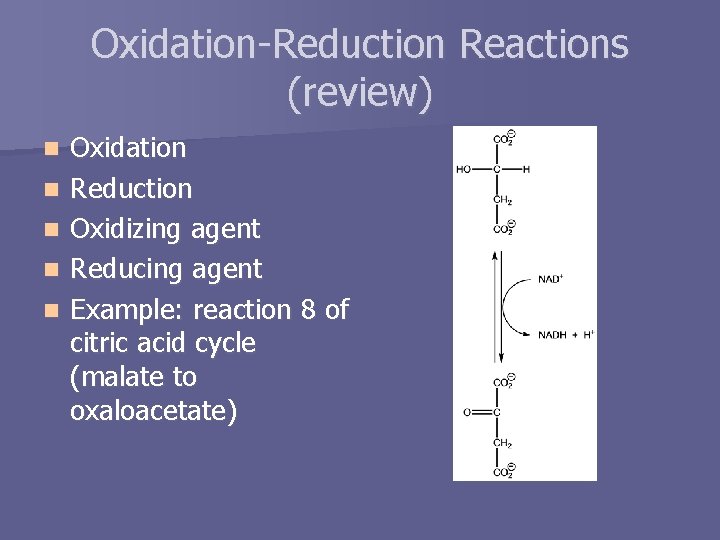
Oxidation-Reduction Reactions (review) n n n Oxidation Reduction Oxidizing agent Reducing agent Example: reaction 8 of citric acid cycle (malate to oxaloacetate)

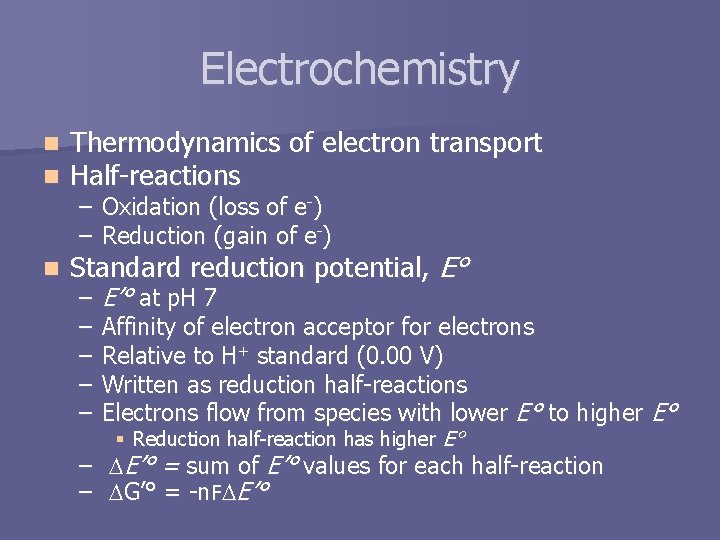
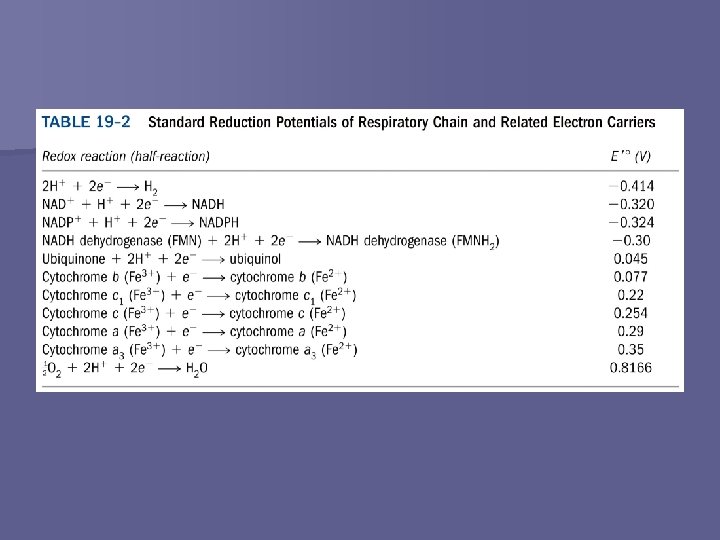
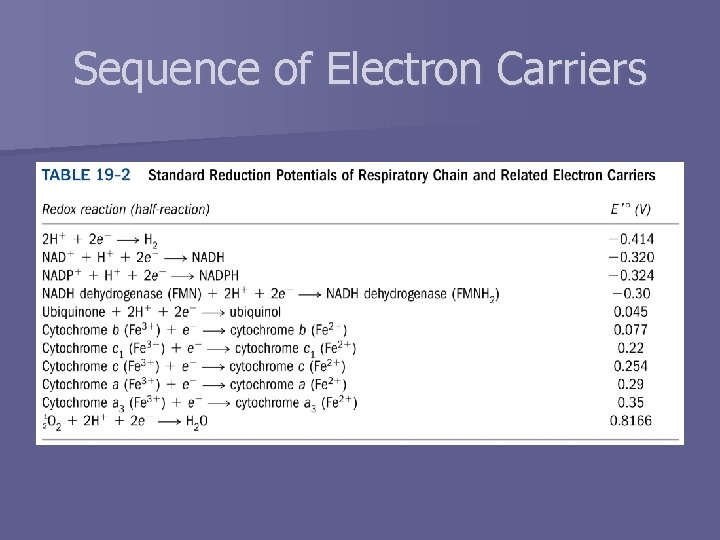
Electrochemistry n n Thermodynamics of electron transport Half-reactions n Standard reduction potential, E° – Oxidation (loss of e-) – Reduction (gain of e-) – – – E’° at p. H 7 Affinity of electron acceptor for electrons Relative to H+ standard (0. 00 V) Written as reduction half-reactions Electrons flow from species with lower E° to higher E° § Reduction half-reaction has higher E° – E’° = sum of E’° values for each half-reaction – G’° = -n. F -n E’°

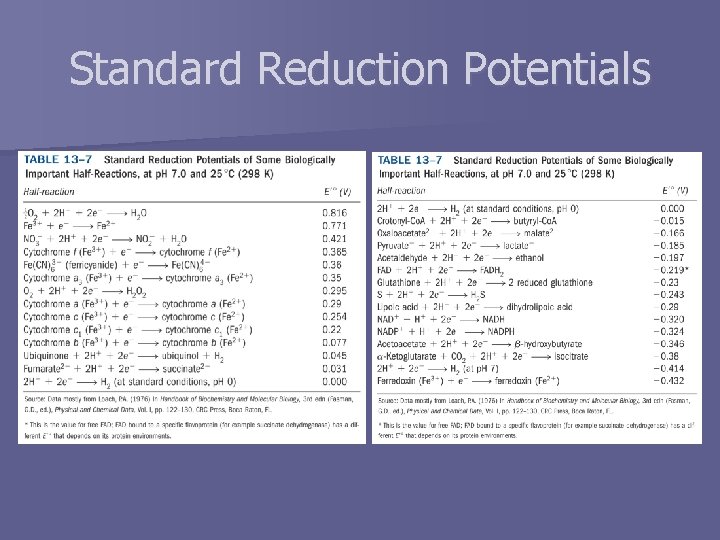
Standard Reduction Potentials

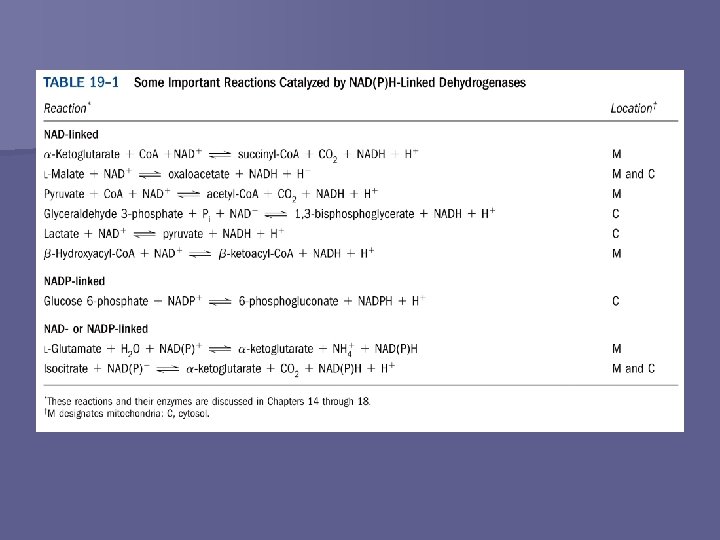
Universal Electron Acceptors n Act as electron carriers – In the form of H atoms (H+ + e-) or hydride ions (H+ + 2 e-) Water-soluble n Undergo reversible oxidation and reduction n Function catalytically (regenerated) n Reduced substrate + NAD+ → oxidized substrate + NADH + H+ n n n NADP+ → NADPH + H+ FAD → FADH 2 FMN → FMNH 2 Q → QH 2 Iron-containing proteins

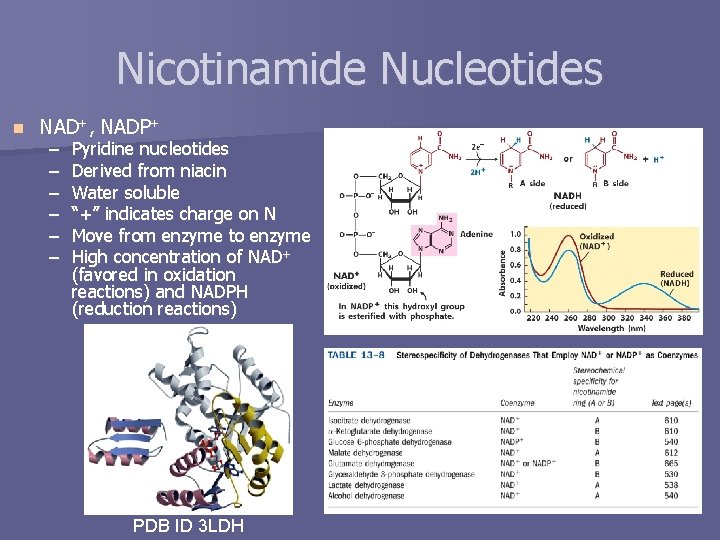
Nicotinamide Nucleotides n NAD+, NADP+ – – – Pyridine nucleotides Derived from niacin Water soluble “+” indicates charge on N Move from enzyme to enzyme High concentration of NAD+ (favored in oxidation reactions) and NADPH (reduction reactions) PDB ID 3 LDH


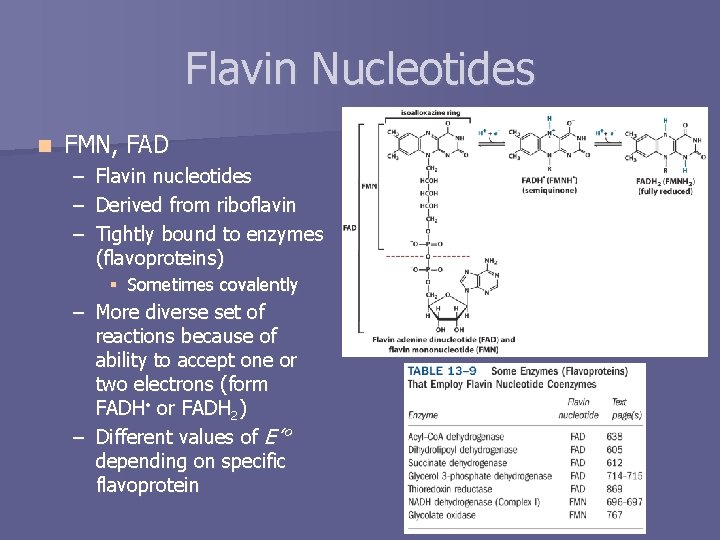
Flavin Nucleotides n FMN, FAD – – – Flavin nucleotides Derived from riboflavin Tightly bound to enzymes (flavoproteins) § Sometimes covalently – More diverse set of reactions because of ability to accept one or two electrons (form FADH • or FADH 2) – Different values of E’° depending on specific flavoprotein

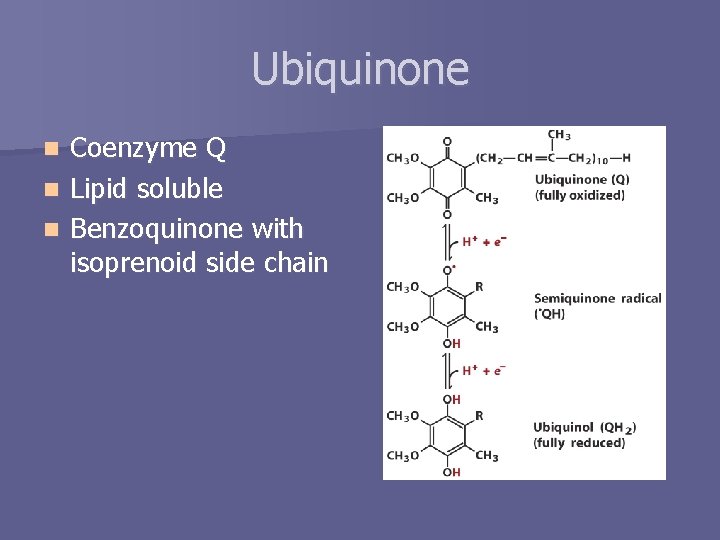
Ubiquinone Coenzyme Q n Lipid soluble n Benzoquinone with isoprenoid side chain n

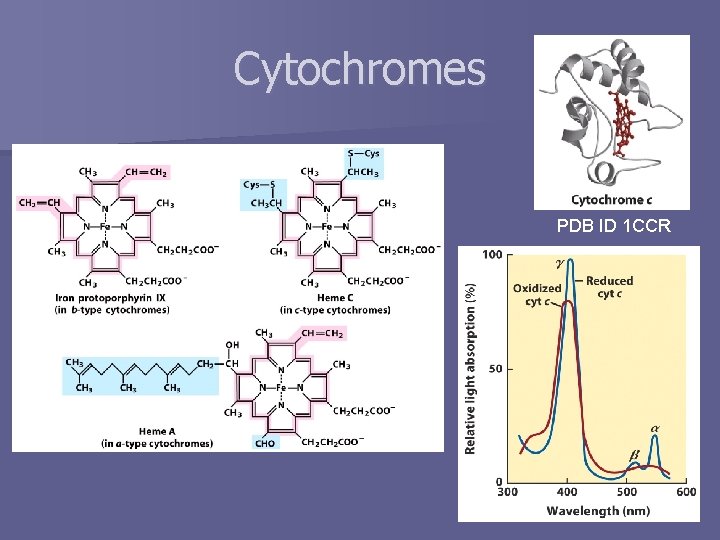
Cytochromes PDB ID 1 CCR


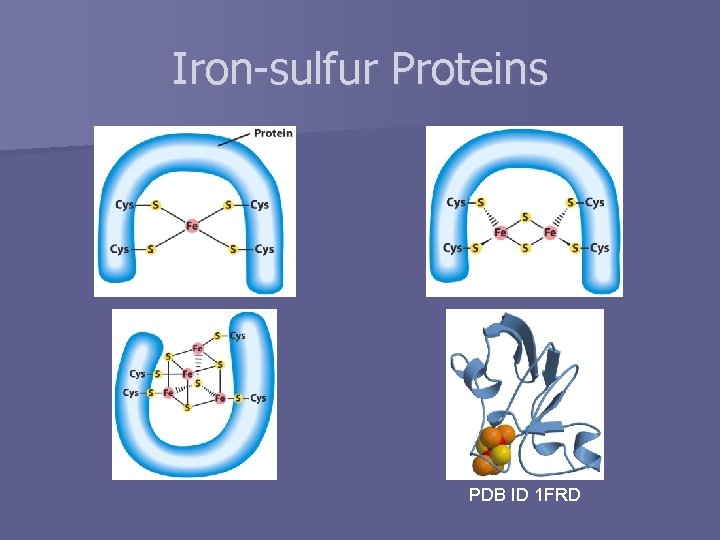
Iron-sulfur Proteins PDB ID 1 FRD

Sequence of Electron Carriers

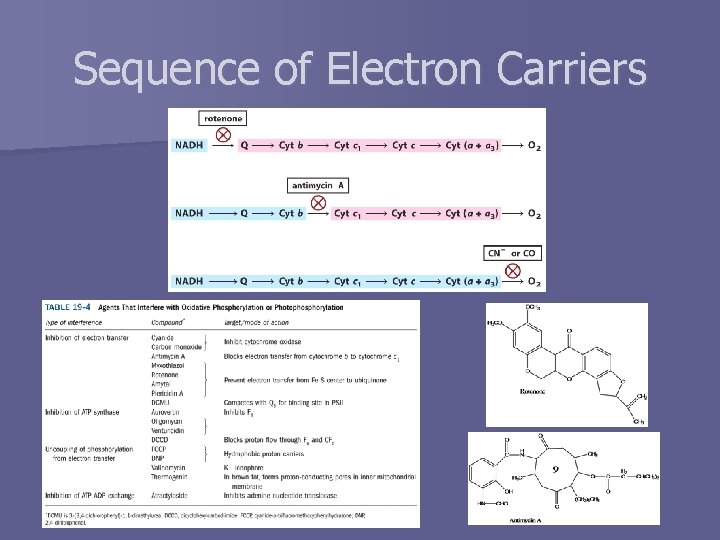
Sequence of Electron Carriers

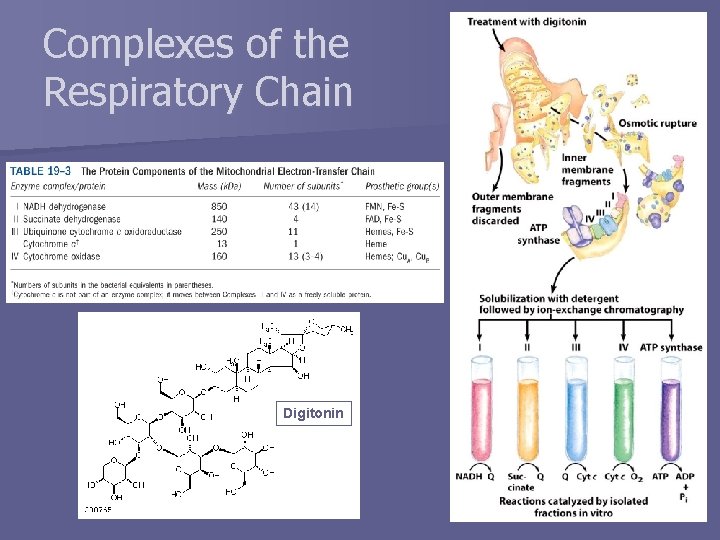
Complexes of the Respiratory Chain Digitonin

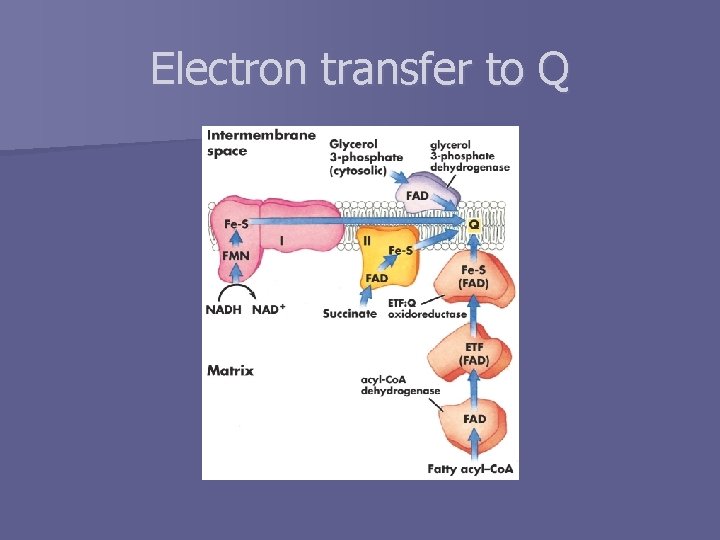
Electron transfer to Q

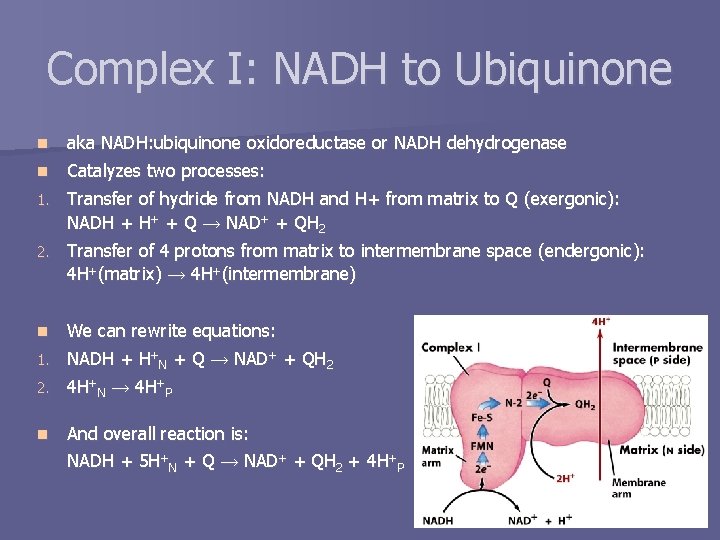
Complex I: NADH to Ubiquinone n aka NADH: ubiquinone oxidoreductase or NADH dehydrogenase n Catalyzes two processes: 1. Transfer of hydride from NADH and H+ from matrix to Q (exergonic): NADH + H+ + Q → NAD+ + QH 2 2. Transfer of 4 protons from matrix to intermembrane space (endergonic): 4 H+(matrix) → 4 H+(intermembrane) We can rewrite equations: 1. NADH + H+N + Q → NAD+ + QH 2 n 2. 4 H+N → 4 H+P n And overall reaction is: NADH + 5 H+N + Q → NAD+ + QH 2 + 4 H+P

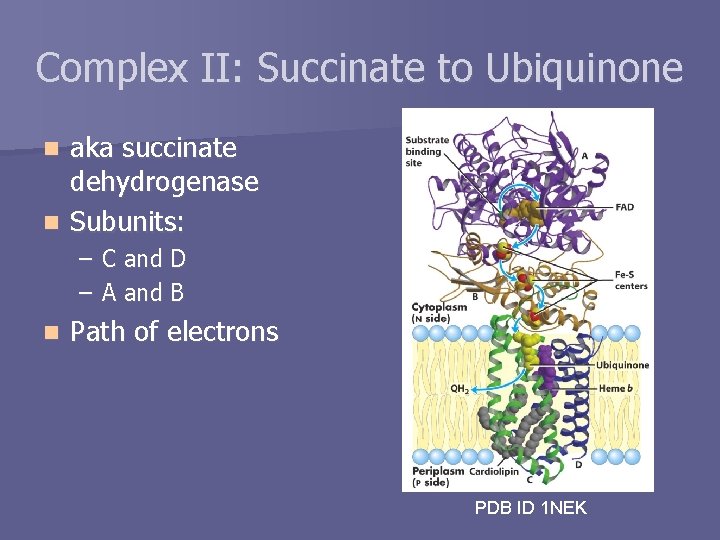
Complex II: Succinate to Ubiquinone aka succinate dehydrogenase n Subunits: n – C and D – A and B n Path of electrons PDB ID 1 NEK

6. Oxidation of succinate to fumarate n Dehydrogenation (loss of H 2; oxidation) n n Catalyzed by succinate dehydrogenase complex n n FADH 2 produced is re-oxidized by coenzyme ubiquinone (Q) to reform FAD and ubiquinol (QH 2) Competitive inhibitor = malonate n -O n Binds to active site through carboxylate groups Cannot undergo dehydrogenation Inhibition reactions used by Krebs to determine citric acid cycle reaction sequence n n n aka succinate dehydrogenase aka Complex II Embedded in inner mitochondrial membrane, rather than in mitochondrial matrix Oxidation of alkane requires stronger oxidizing agent than NAD+ (hence FAD) n n Stereospecific to form trans double bond only 2 C-CH 2 -CO 2 Symmetrical molecule evenly distributes carbons in remainder of products throughout the cycle

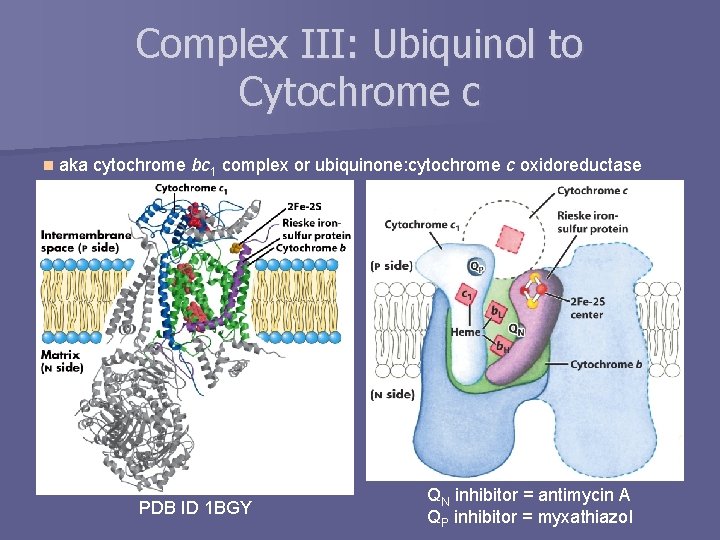
Complex III: Ubiquinol to Cytochrome c n aka cytochrome bc 1 complex or ubiquinone: cytochrome c oxidoreductase PDB ID 1 BGY QN inhibitor = antimycin A QP inhibitor = myxathiazol

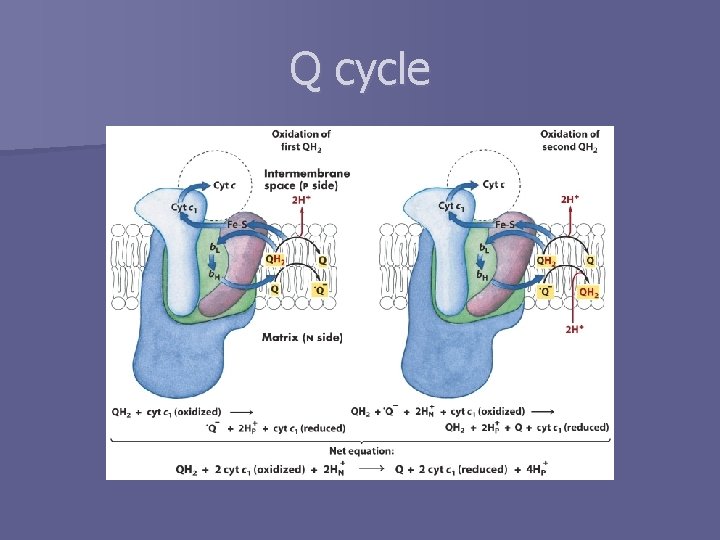
Q cycle

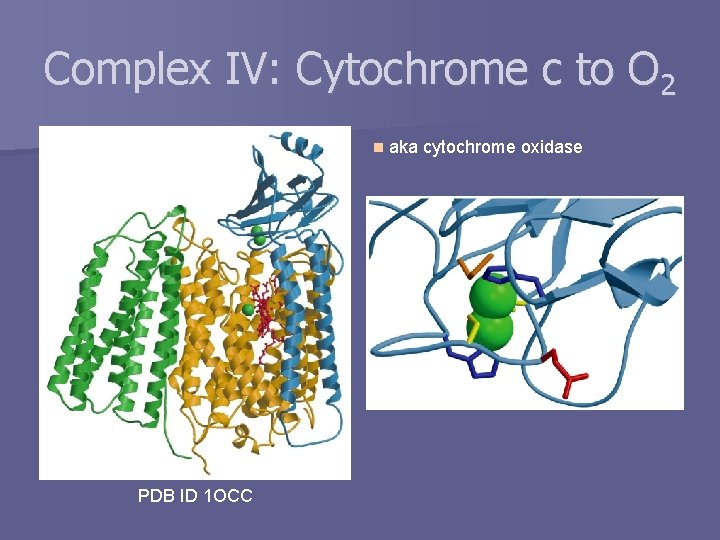
Complex IV: Cytochrome c to O 2 n PDB ID 1 OCC aka cytochrome oxidase

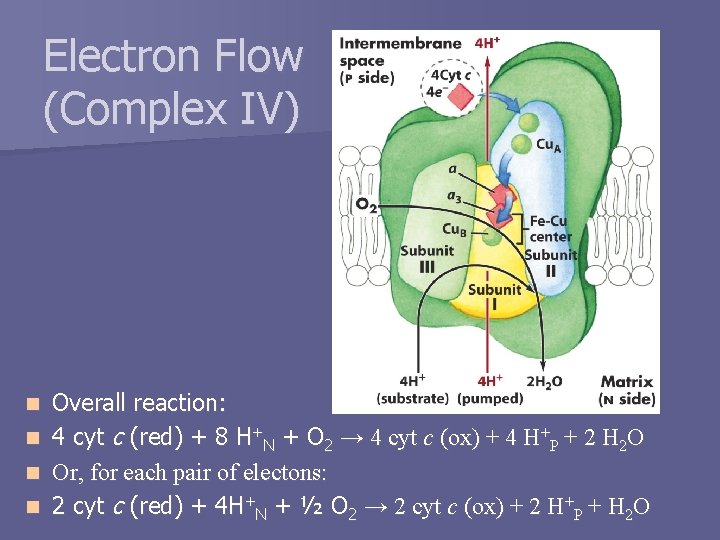
Electron Flow (Complex IV) n n Overall reaction: 4 cyt c (red) + 8 H+N + O 2 → 4 cyt c (ox) + 4 H+P + 2 H 2 O Or, for each pair of electons: 2 cyt c (red) + 4 H+N + ½ O 2 → 2 cyt c (ox) + 2 H+P + H 2 O

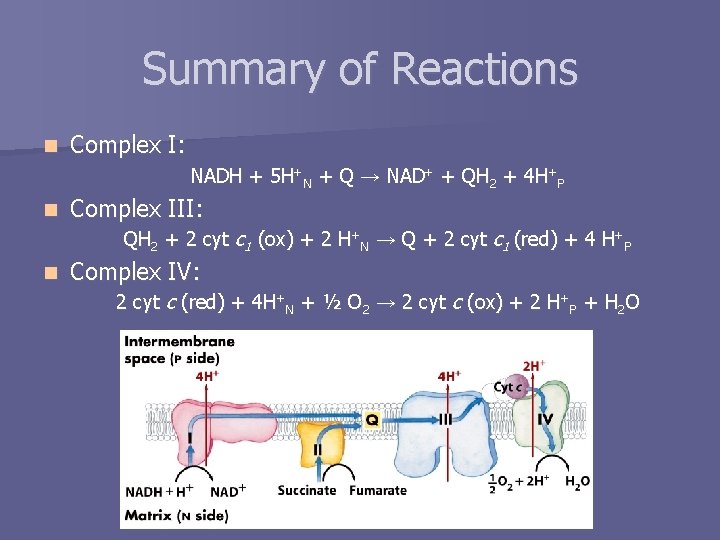
Summary of Reactions n Complex I: NADH + 5 H+N + Q → NAD+ + QH 2 + 4 H+P n Complex III: QH 2 + 2 cyt c 1 (ox) + 2 H+N → Q + 2 cyt c 1 (red) + 4 H+P n Complex IV: 2 cyt c (red) + 4 H+N + ½ O 2 → 2 cyt c (ox) + 2 H+P + H 2 O

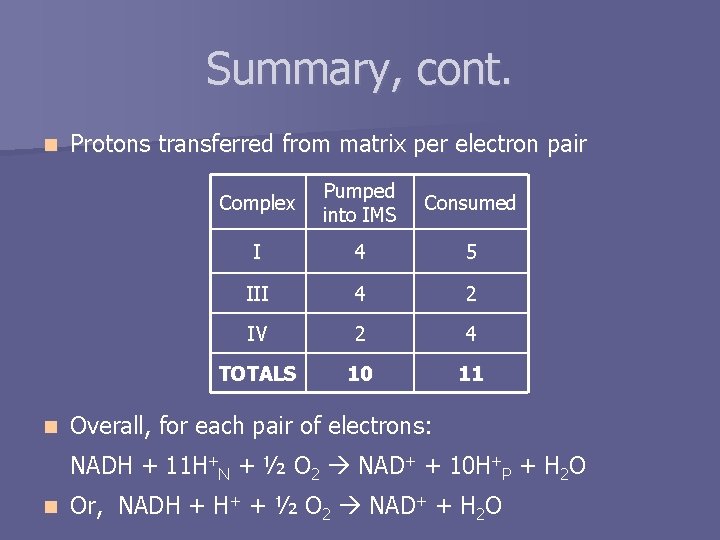
Summary, cont. n n Protons transferred from matrix per electron pair Complex Pumped into IMS Consumed I 4 5 III 4 2 IV 2 4 TOTALS 10 11 Overall, for each pair of electrons: NADH + 11 H+N + ½ O 2 NAD+ + 10 H+P + H 2 O n Or, NADH + H+ + ½ O 2 NAD+ + H 2 O

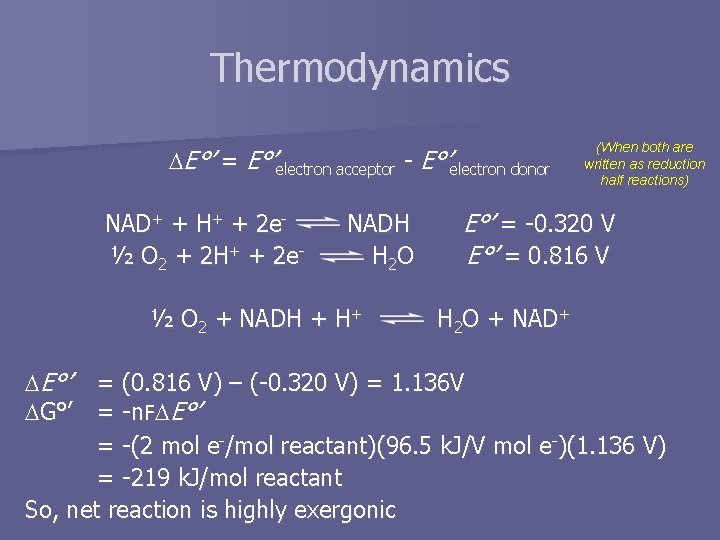
Thermodynamics E°’ = E°’electron acceptor - E°’electron donor NAD+ + H+ + 2 e½ O 2 + 2 H+ + 2 e- NADH H 2 O ½ O 2 + NADH + H+ (When both are written as reduction half reactions) E°’ = -0. 320 V E°’ = 0. 816 V H 2 O + NAD+ E°’ = (0. 816 V) – (-0. 320 V) = 1. 136 V G°’ = -n. F -n E°’ = -(2 mol e-/mol reactant)(96. 5 k. J/V mol e-)(1. 136 V) = -219 k. J/mol reactant So, net reaction is highly exergonic

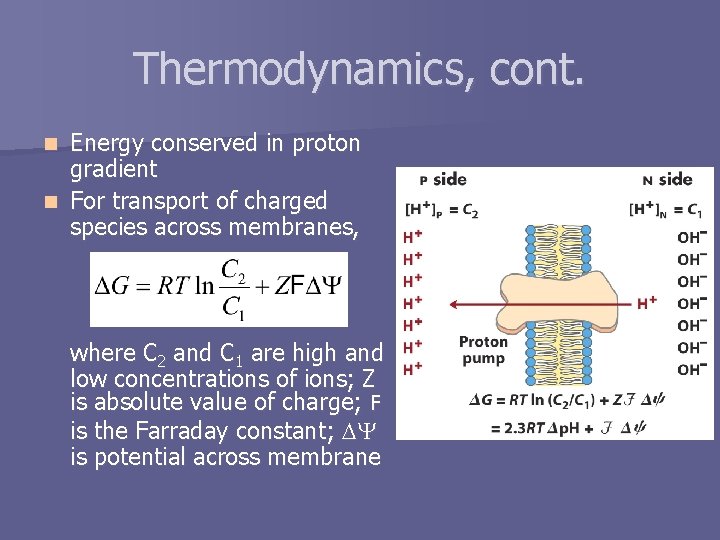
Thermodynamics, cont. Energy conserved in proton gradient n For transport of charged species across membranes, n where C 2 and C 1 are high and low concentrations of ions; Z is absolute value of charge; F is the Farraday constant; is potential across membrane

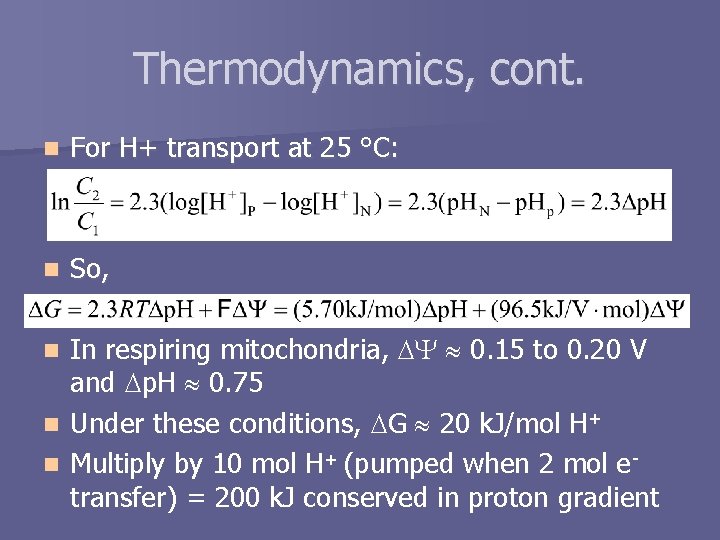
Thermodynamics, cont. n For H+ transport at 25 °C: n So, In respiring mitochondria, 0. 15 to 0. 20 V and p. H 0. 75 n Under these conditions, G 20 k. J/mol H+ n Multiply by 10 mol H+ (pumped when 2 mol etransfer) = 200 k. J conserved in proton gradient n

Next… n Synthesis of ATP

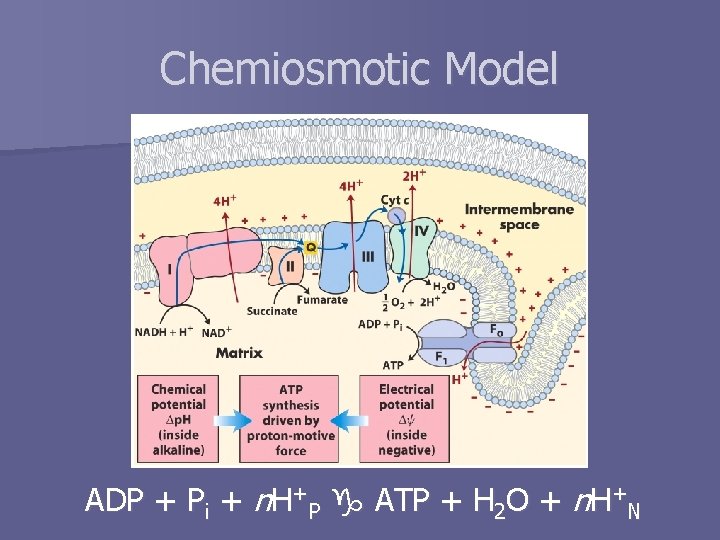
ATP Synthesis n Energy from electron transfer reactions conserved in proton gradient – Chemical (concentration) – Electrical (charge) n This energy is used to drive ATP synthesis n Chemiosmotic model

Chemiosmotic Model ADP + Pi + n. H+P ATP + H 2 O + n. H+N

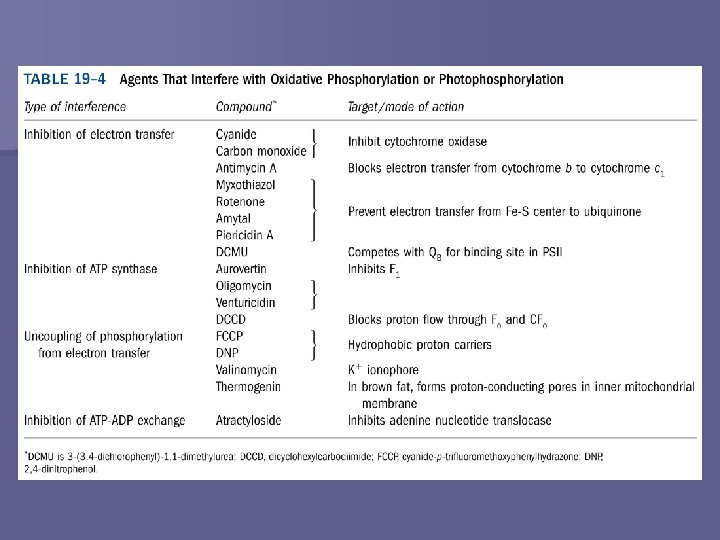
Coupling of Electron Transfer and ATP Synthesis Table 19 -4

![Experimental Support K Cl Experimental Support [K+] < [Cl-]](https://slidetodoc.com/presentation_image_h2/9d0187dabf181bb00c67f00554c21b3e/image-36.jpg)
Experimental Support [K+] < [Cl-]

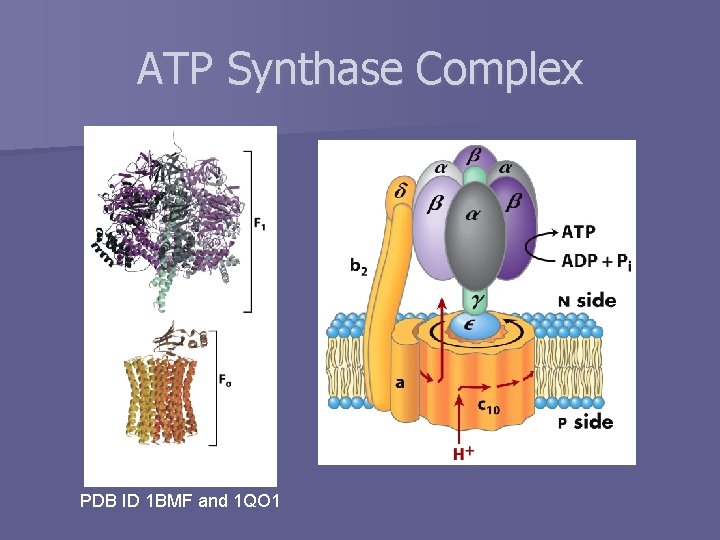
ATP Synthase Complex PDB ID 1 BMF and 1 QO 1

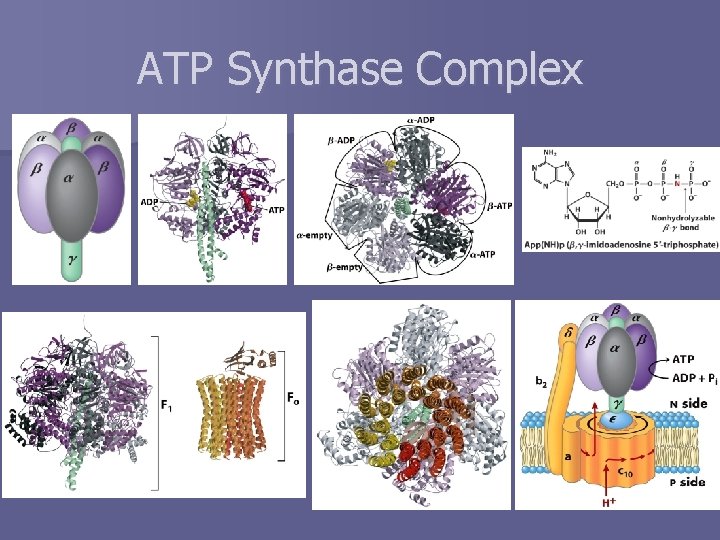
ATP Synthase Complex

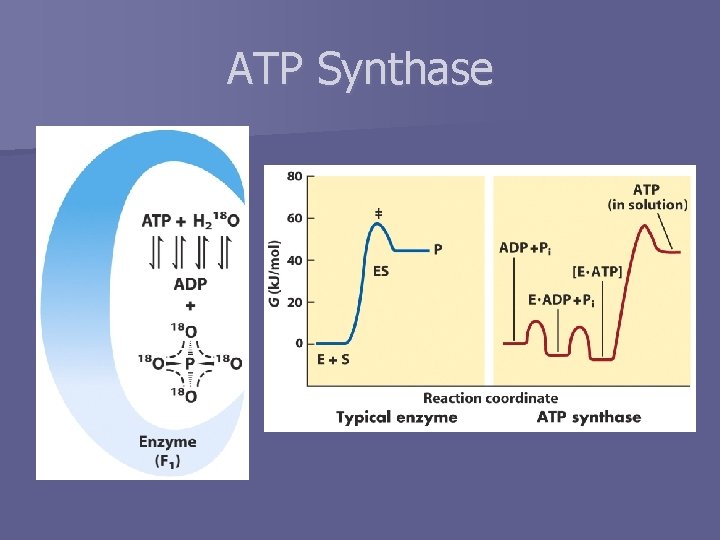
ATP Synthase

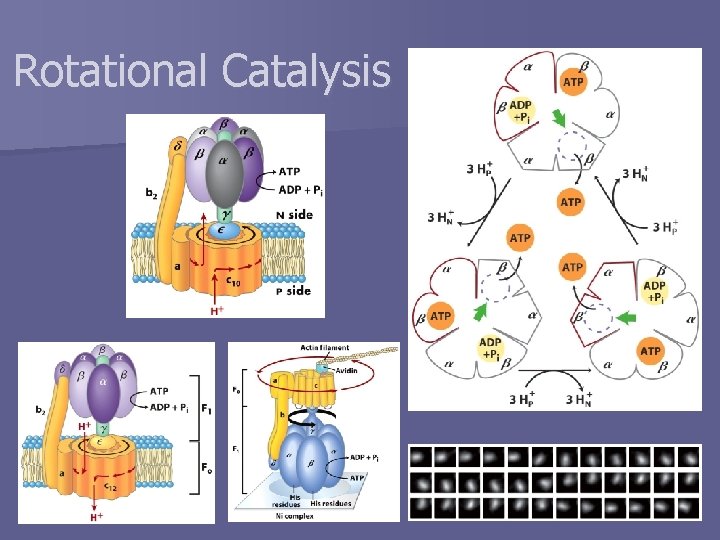
Rotational Catalysis

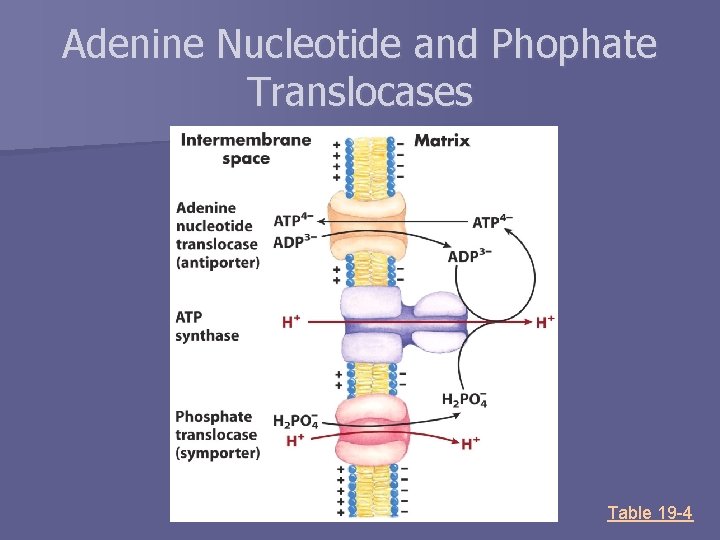
Adenine Nucleotide and Phophate Translocases Table 19 -4

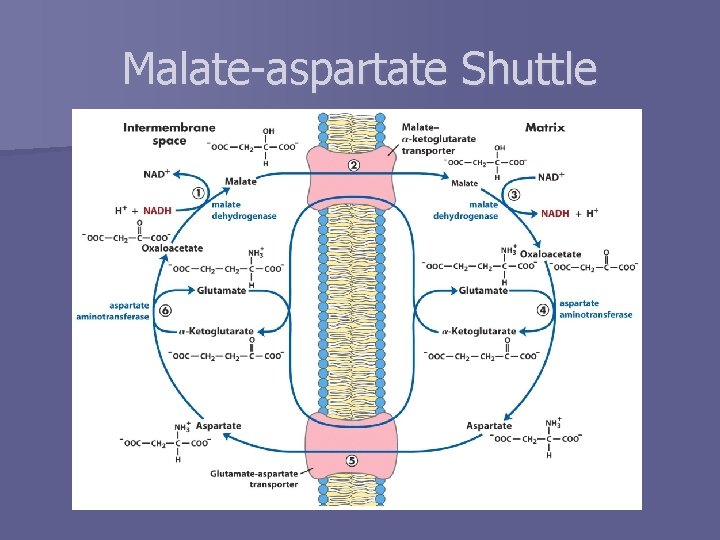
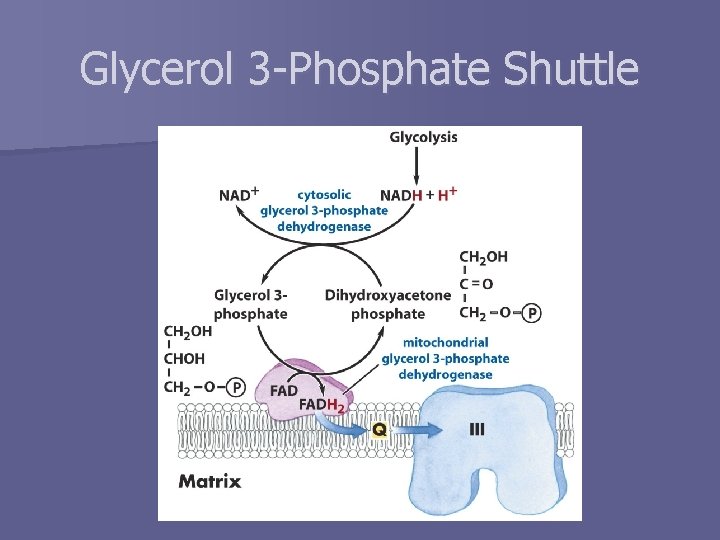
How do electrons get into the matrix? n Inner mitochondrial membrane is impermeable to NADH n Two possible methods to transfer reducing equivalents “through” membrane – Liver, kidney, heart § Electrons are instead transferred to malate § Malate-aspartate shuttle – Skeletal muscle, brain § Glycerol 3 -phosphate shuttle

Malate-aspartate Shuttle

Glycerol 3 -Phosphate Shuttle

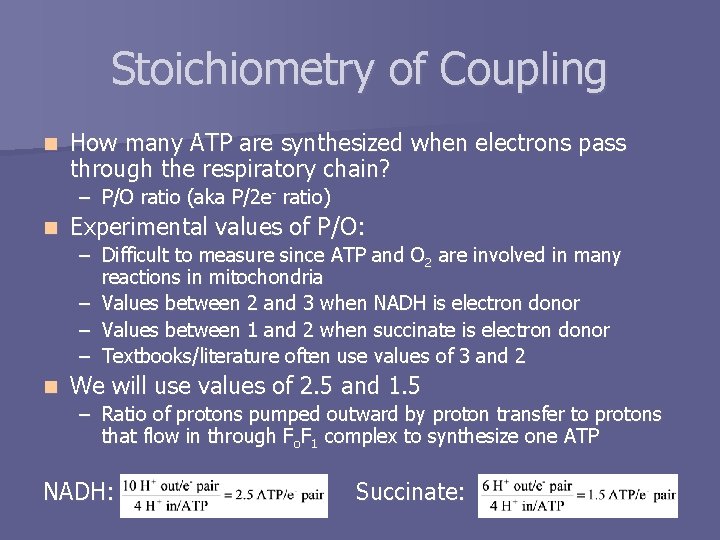
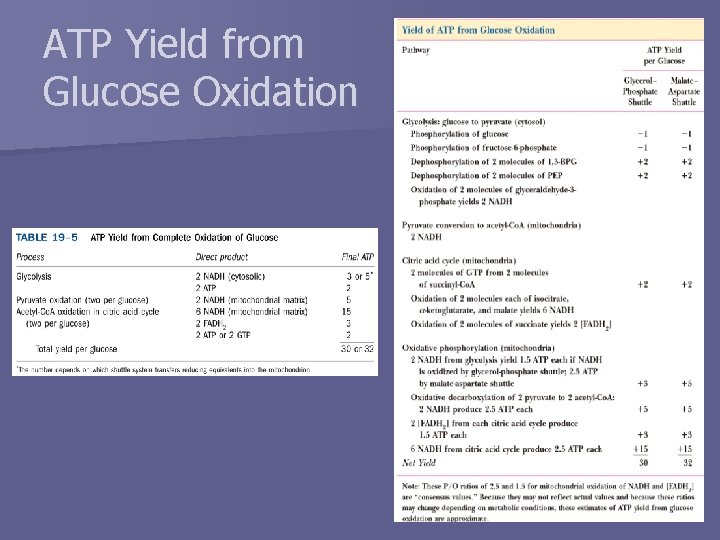
Stoichiometry of Coupling n How many ATP are synthesized when electrons pass through the respiratory chain? – P/O ratio (aka P/2 e- ratio) n Experimental values of P/O: – Difficult to measure since ATP and O 2 are involved in many reactions in mitochondria – Values between 2 and 3 when NADH is electron donor – Values between 1 and 2 when succinate is electron donor – Textbooks/literature often use values of 3 and 2 n We will use values of 2. 5 and 1. 5 – Ratio of protons pumped outward by proton transfer to protons that flow in through Fo. F 1 complex to synthesize one ATP NADH: Succinate:

ATP Yield from Glucose Oxidation n How many ATP are produced from the complete oxidation of 1 glucose molecule to CO 2, assuming NADH enters the mitochondrion via the malate-aspartate shuttle? n How many ATP are produced from the complete oxidation of 1 glucose molecule to CO 2, assuming NADH enters the mitochondrion via the glycerol 3 -phosphate shuttle?

ATP Yield from Glucose Oxidation

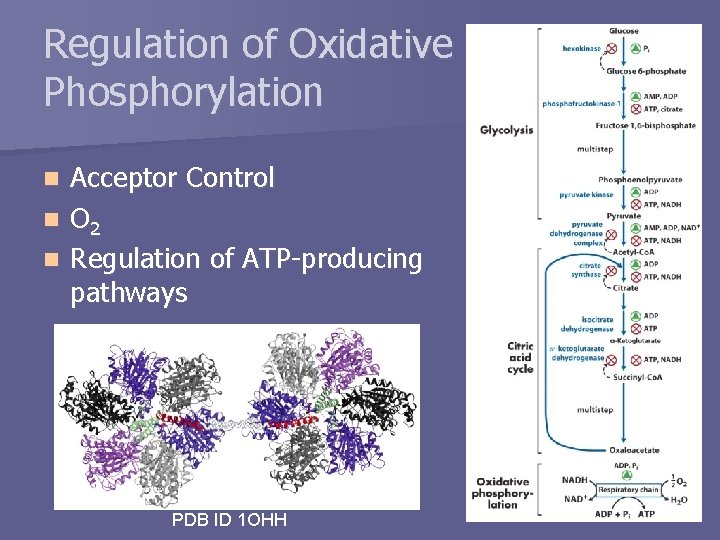
Regulation of Oxidative Phosphorylation Acceptor Control n O 2 n Regulation of ATP-producing pathways n PDB ID 1 OHH