Overview of SNP Genotyping Debbie Nickerson Department of

Overview of SNP Genotyping Debbie Nickerson Department of Genome Sciences University of Washington debnick@u. washington. edu

SNP Genotyping - Overview • Project Rationale • Genotyping Strategies/Technical Leaps • Data Management/Quality Control
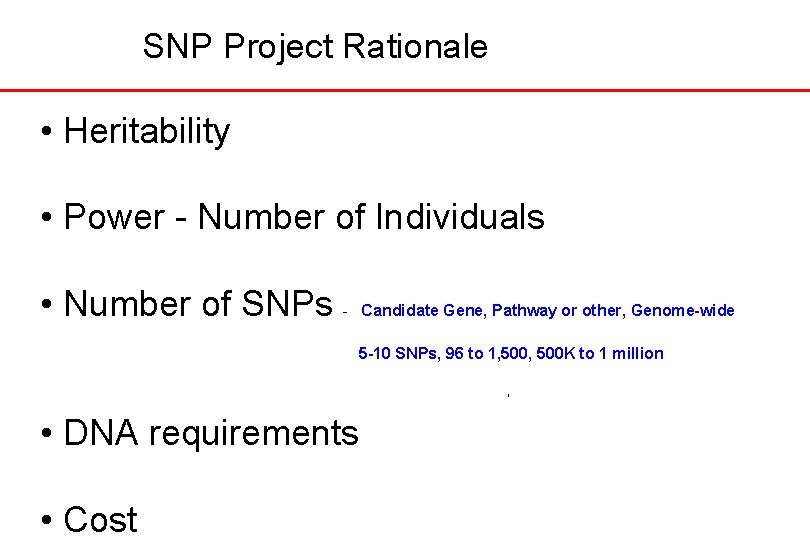
SNP Project Rationale • Heritability • Power - Number of Individuals • Number of SNPs - Candidate Gene, Pathway or other, Genome-wide 5 -10 SNPs, 96 to 1, 500 K to 1 million • DNA requirements • Cost
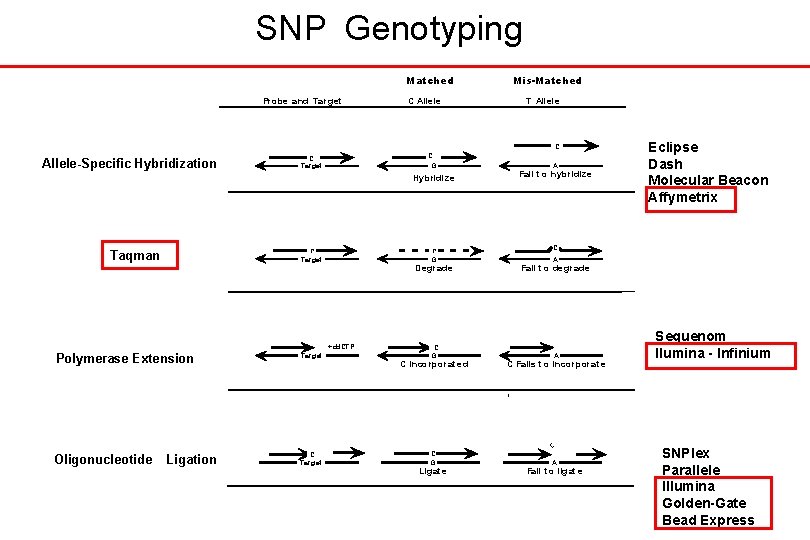
SNP Genotyping Matched Probe and Target C Allele Mis-Matched T Allele C Allele-Specific Hybridization C G C Target Hyb ridiz e C Target Taqman C G Deg rade +dd. CTP Polymerase Extension Target Oligonucleotide C Target Ligation C G C inco rporat ed C G Ligat e A Fail t o hybridize Eclipse Dash Molecular Beacon Affymetrix C A Fail t o deg rade A C Fails t o incorporat e C A Fail t o ligat e Sequenom Ilumina - Infinium SNPlex Parallele Illumina Golden-Gate Bead Express
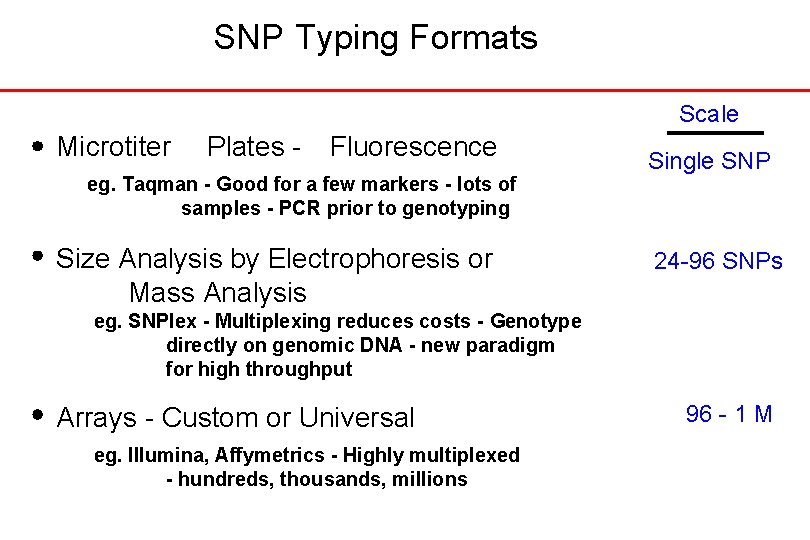
SNP Typing Formats Scale Microtiter Plates - Fluorescence eg. Taqman - Good for a few markers - lots of samples - PCR prior to genotyping Size Analysis by Electrophoresis or Mass Analysis Single SNP 24 -96 SNPs eg. SNPlex - Multiplexing reduces costs - Genotype directly on genomic DNA - new paradigm for high throughput Arrays - Custom or Universal eg. Illumina, Affymetrics - Highly multiplexed - hundreds, thousands, millions 96 - 1 M
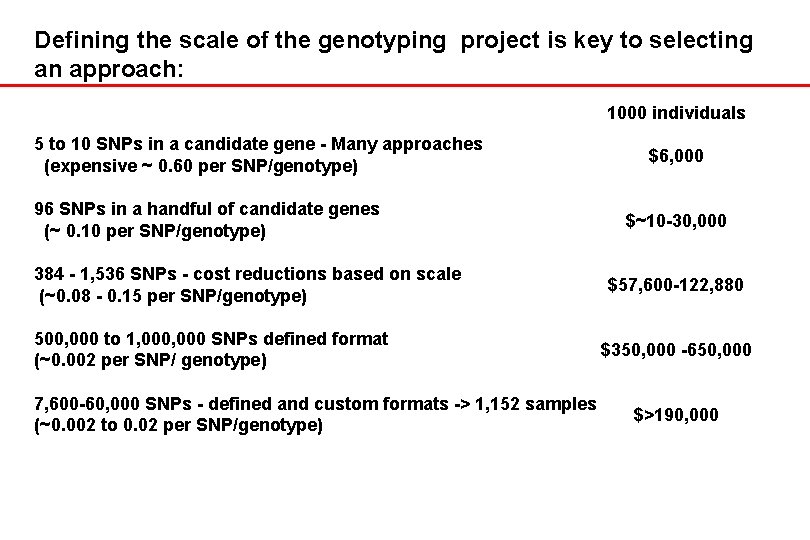
Defining the scale of the genotyping project is key to selecting an approach: 1000 individuals 5 to 10 SNPs in a candidate gene - Many approaches (expensive ~ 0. 60 per SNP/genotype) 96 SNPs in a handful of candidate genes (~ 0. 10 per SNP/genotype) 384 - 1, 536 SNPs - cost reductions based on scale (~0. 08 - 0. 15 per SNP/genotype) 500, 000 to 1, 000 SNPs defined format (~0. 002 per SNP/ genotype) 7, 600 -60, 000 SNPs - defined and custom formats -> 1, 152 samples (~0. 002 to 0. 02 per SNP/genotype) $6, 000 $~10 -30, 000 $57, 600 -122, 880 $350, 000 -650, 000 $>190, 000
Many Approaches to Genotype a Handful of SNPs PCR region prior to SNP genotyping - Adds to cost - Many use modified primers - the more modified, the higher the cost • Taqman * • Single base extension - Sequenom - Mass Spec Illumina - Infinium • Eclipse • Dash • Molecular Beacons
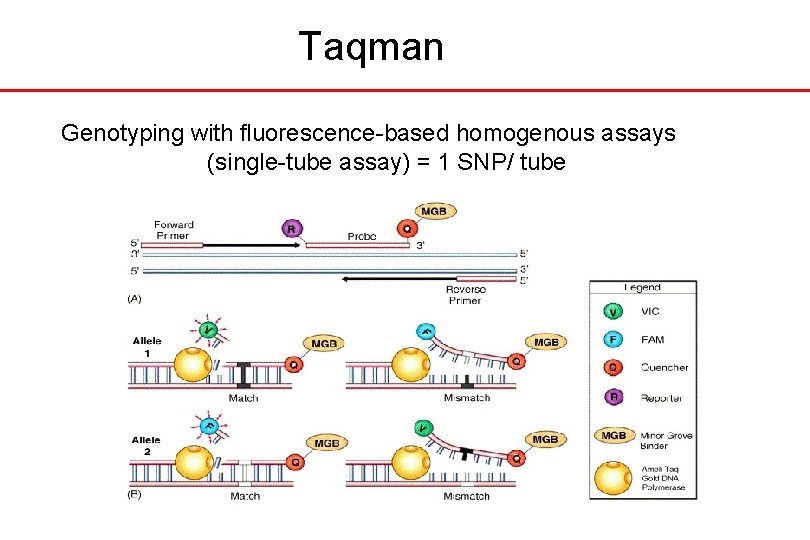
Taqman Genotyping with fluorescence-based homogenous assays (single-tube assay) = 1 SNP/ tube
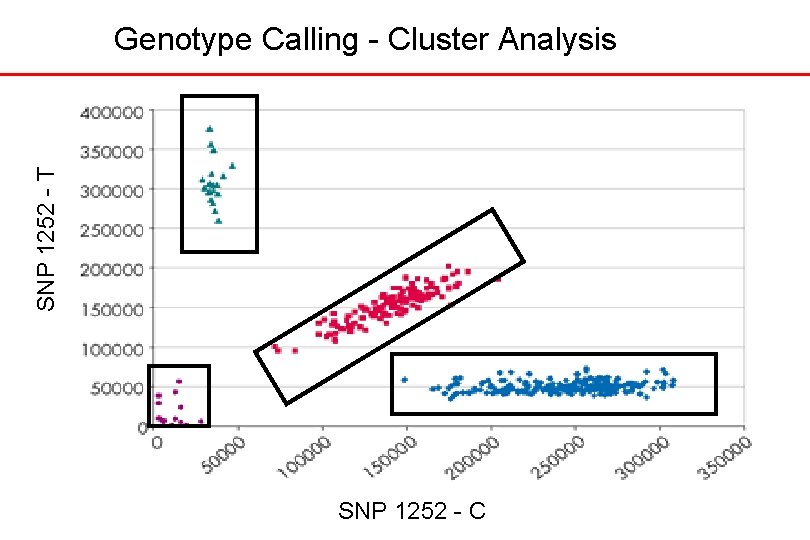
SNP 1252 - T Genotype Calling - Cluster Analysis SNP 1252 - C
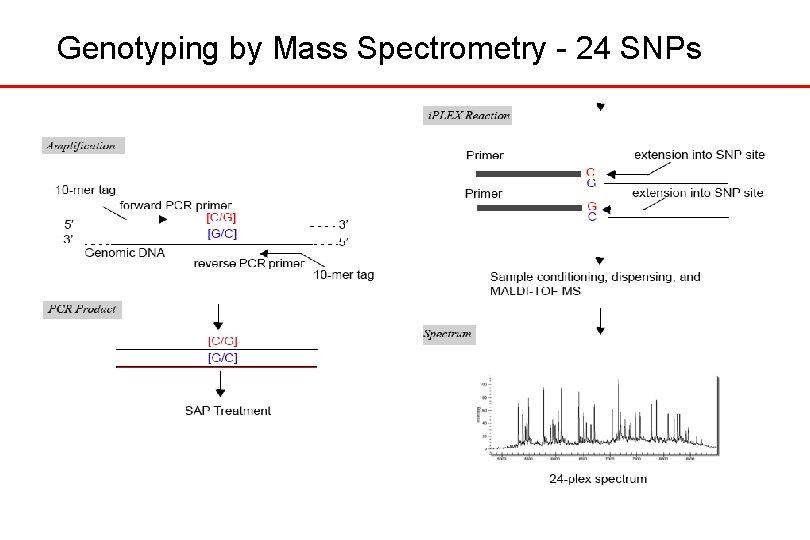
Genotyping by Mass Spectrometry - 24 SNPs

Technological Leap - No advance PCR Universal PCR after preparing multiple regions for analysis Several based on primer specific on genomic DNA followed by PCR of the ligated products - different strategies and different readouts. SNPlex (ABI), Illumina (Bead Express, Golden-Gate), Affymetrix Also, Genome-wide: Reduced representation - Affymetrix Whole Genome Amplification - Illumina
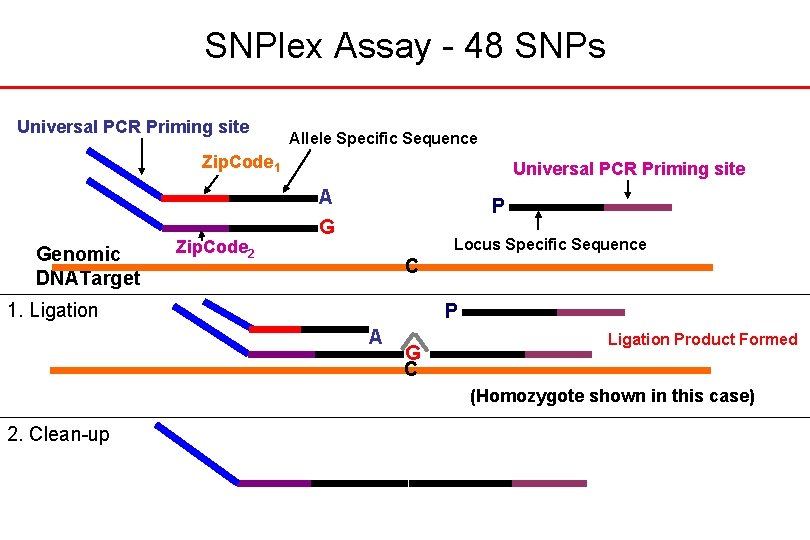
SNPlex Assay - 48 SNPs Universal PCR Priming site Allele Specific Sequence Zip. Code 1 Genomic DNATarget Zip. Code 2 Universal PCR Priming site A G P Locus Specific Sequence C 1. Ligation P A G C Ligation Product Formed (Homozygote shown in this case) 2. Clean-up
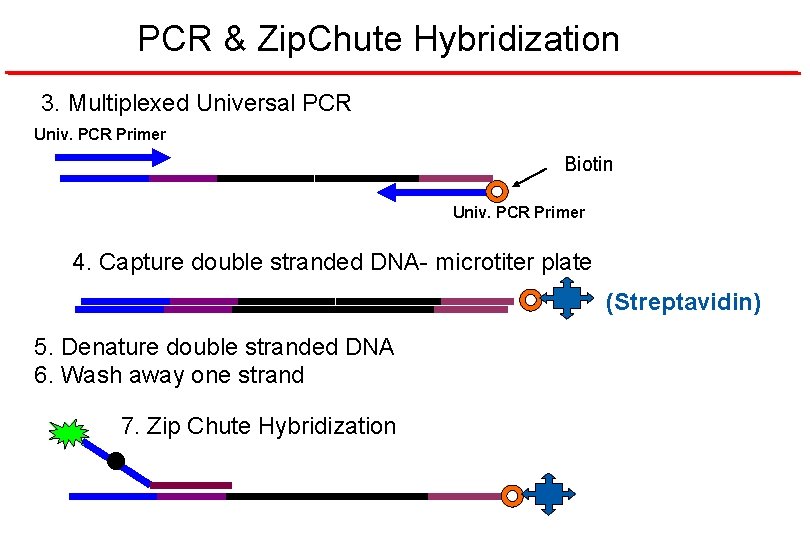
PCR & Zip. Chute Hybridization 3. Multiplexed Universal PCR Univ. PCR Primer Biotin Univ. PCR Primer 4. Capture double stranded DNA- microtiter plate (Streptavidin) 5. Denature double stranded DNA 6. Wash away one strand 7. Zip Chute Hybridization •
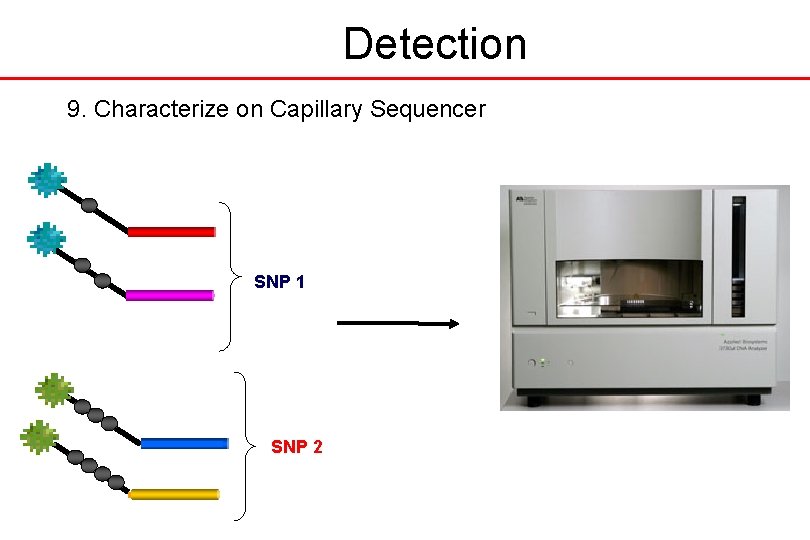
Detection 9. Characterize on Capillary Sequencer SNP 1 SNP 2
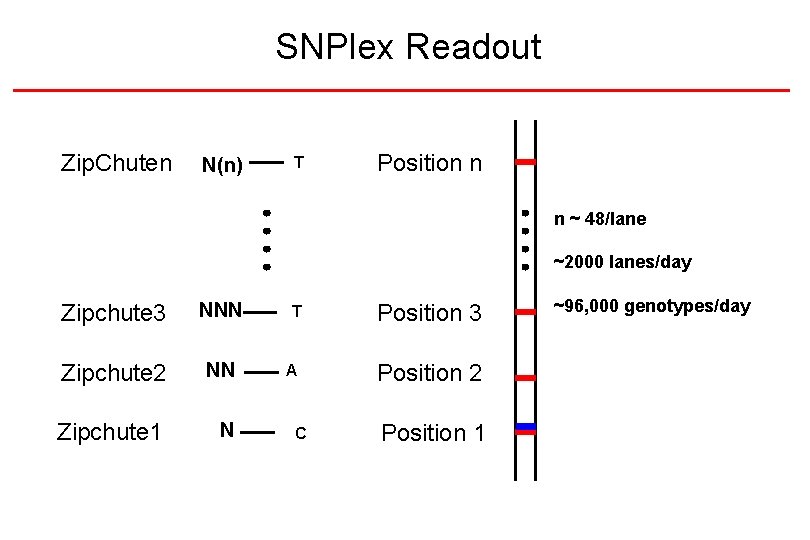
SNPlex Readout Zip. Chuten N(n) T Position n n ~ 48/lane ~2000 lanes/day Zipchute 3 NNN T Position 3 Zipchute 2 NN A Position 2 Zipchute 1 N C Position 1 ~96, 000 genotypes/day
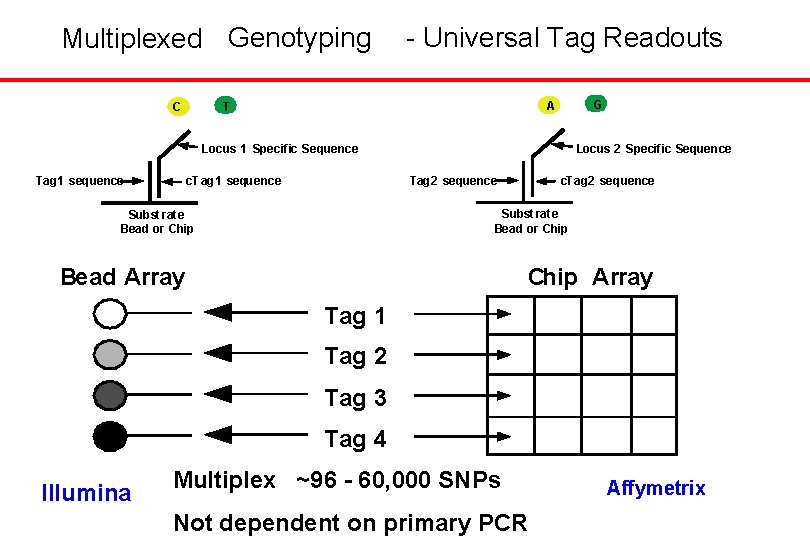
Multiplexed Genotyping C - Universal Tag Readouts G A T Locus 2 Specif ic Sequence Locus 1 Specific Sequence Tag 1 sequence Tag 2 sequence c. Tag 1 sequence c. Tag 2 sequence Subst rat e Bead or Chip Bead Array Chip Array Tag 1 Tag 2 Tag 3 Tag 4 Illumina Multiplex ~96 - 60, 000 SNPs Not dependent on primary PCR Affymetrix
Arrays - High Density Genotyping Thousands of SNPs and Beyond • “Bead” Arrays - Illumina – Manufactured by self-assembly – Beads identified by decoding
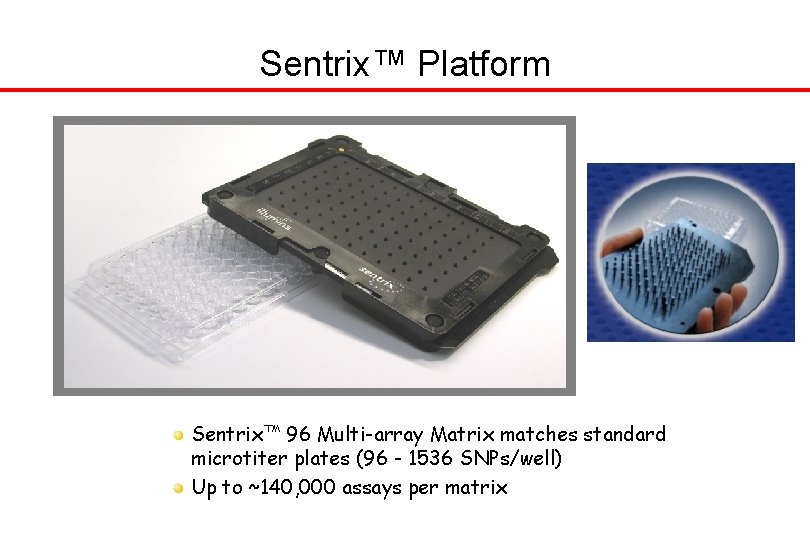
Sentrix™ Platform Sentrix™ 96 Multi-array Matrix matches standard microtiter plates (96 - 1536 SNPs/well) Up to ~140, 000 assays per matrix
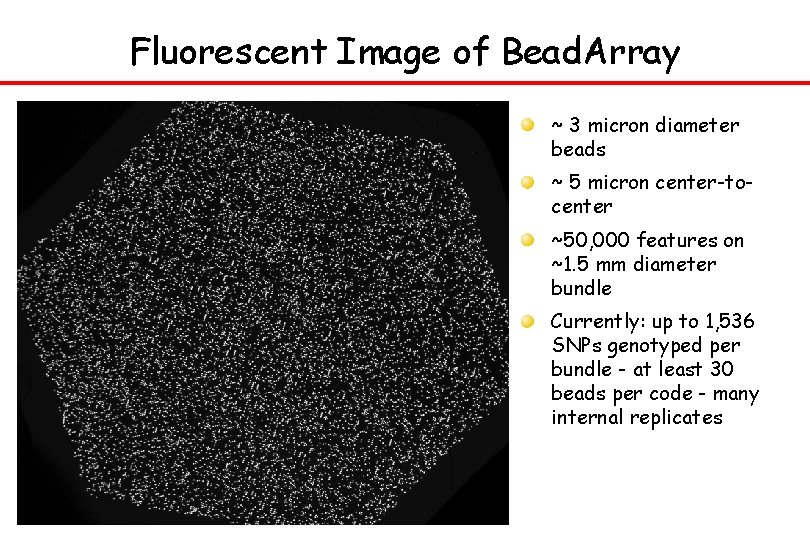
Fluorescent Image of Bead. Array ~ 3 micron diameter beads ~ 5 micron center-tocenter ~50, 000 features on ~1. 5 mm diameter bundle Currently: up to 1, 536 SNPs genotyped per bundle - at least 30 beads per code - many internal replicates
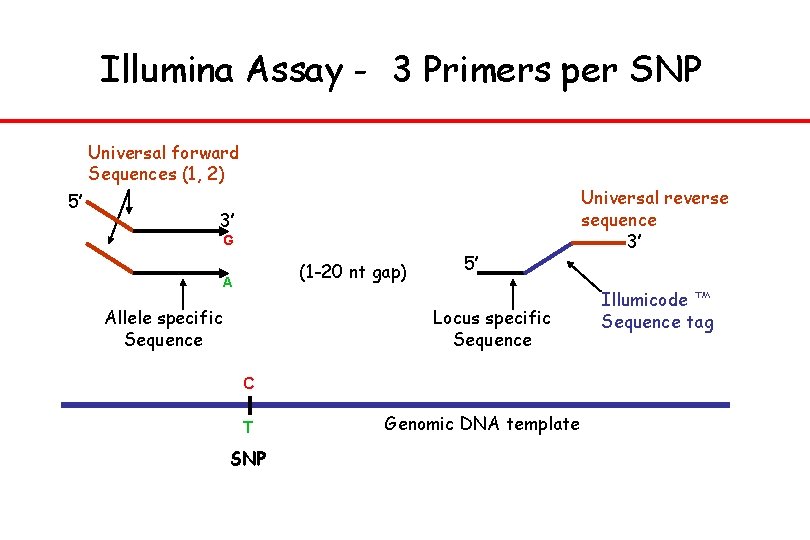
Illumina Assay - 3 Primers per SNP Universal forward Sequences (1, 2) 5’ 3’ G (1 -20 nt gap) A Allele specific Sequence 5’ Locus specific Sequence C T SNP Genomic DNA template Universal reverse sequence 3’ Illumicode ™ Sequence tag
![Allele-Specific Extension and Ligation Polymerase Genomic DNA Allele Specific Extension & Ligation [T/C] Universal Allele-Specific Extension and Ligation Polymerase Genomic DNA Allele Specific Extension & Ligation [T/C] Universal](http://slidetodoc.com/presentation_image_h2/0ed57e17aaa567c1cf29c52e6179b01c/image-21.jpg)
Allele-Specific Extension and Ligation Polymerase Genomic DNA Allele Specific Extension & Ligation [T/C] Universal PCR Sequence 1 Universal PCR Sequence 2 A G Ligase [T/A] illumi. Code’ Address Universal PCR Sequence 3’
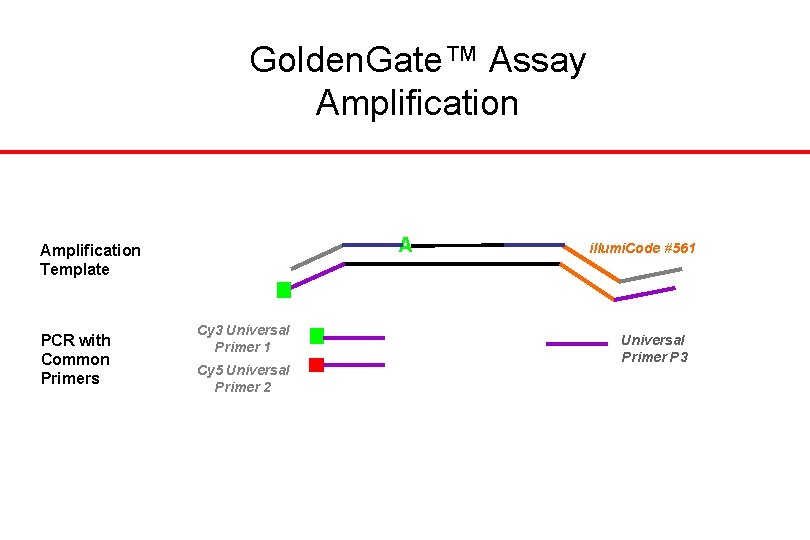
Golden. Gate™ Assay Amplification A Amplification Template PCR with Common Primers Cy 3 Universal Primer 1 Cy 5 Universal Primer 2 illumi. Code #561 Universal Primer P 3
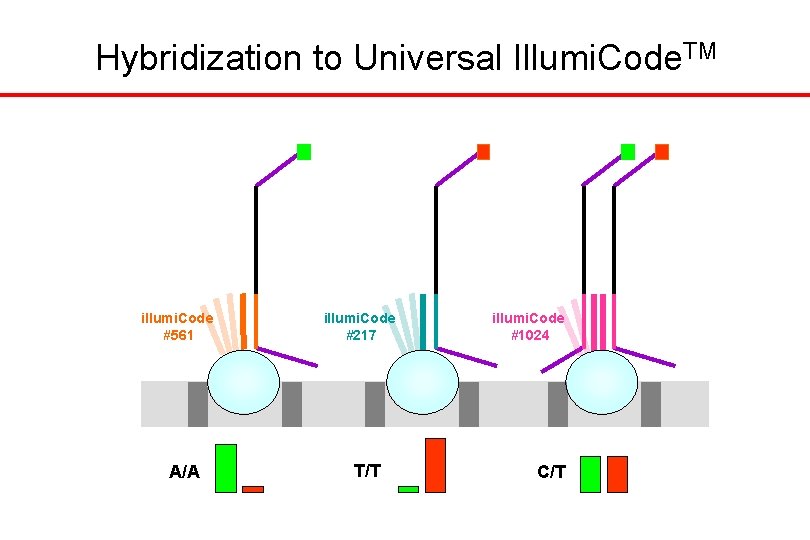
Hybridization to Universal Illumi. Code. TM //// A/A illumi. Code #1024 //// illumi. Code #217 T/T //// illumi. Code #561 C/T
Bead. Array Reader • • • Confocal laser scanning system Resolution, 0. 8 micron Two lasers 532, 635 nm – Supports Cy 3 & Cy 5 imaging • Sentrix Arrays (96 bundle) and Slides for 100 k fixed formats
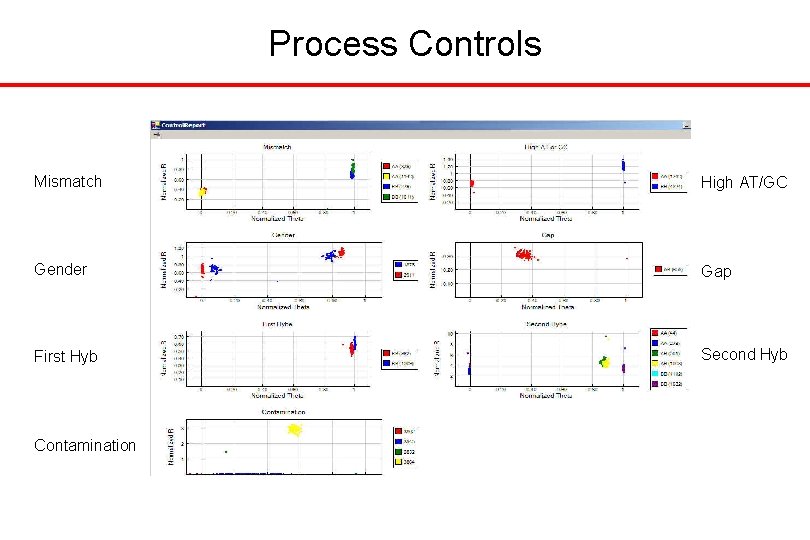
Process Controls Mismatch High AT/GC Gender Gap First Hyb Second Hyb Contamination
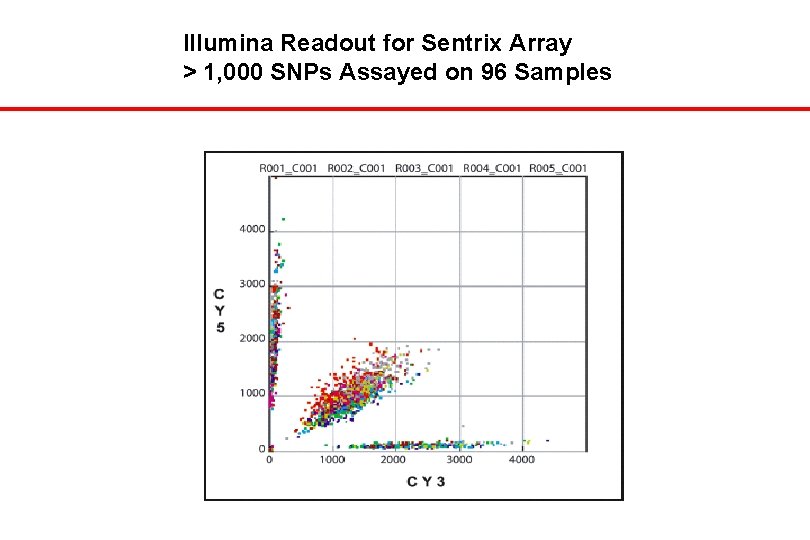
Illumina Readout for Sentrix Array > 1, 000 SNPs Assayed on 96 Samples

Genotyping for Whole Genome Association Studies • Rapid Advances in Whole Genome Platforms • Significant Content Improvements now 1 million SNP chips are the standard • Increasing coverage of multiple populations • Decreasing costs • Some drop in content when populations beyond the Hap. Map are genotyped De. Bakker et al Nat. Genet. 38: 1298, 2006 Conrad et al Nat. Genet. 38: 1251, 2006
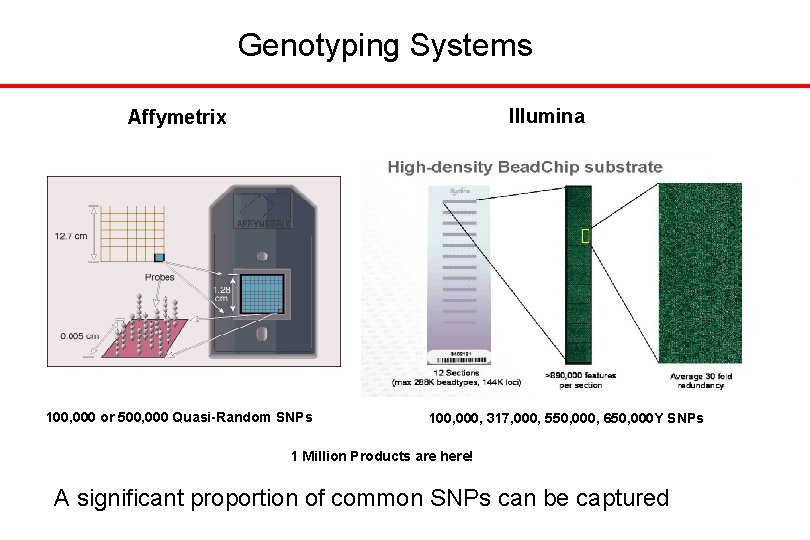
Genotyping Systems Illumina Affymetrix 100, 000 or 500, 000 Quasi-Random SNPs 100, 000, 317, 000, 550, 000, 650, 000 Y SNPs 1 Million Products are here! A significant proportion of common SNPs can be captured
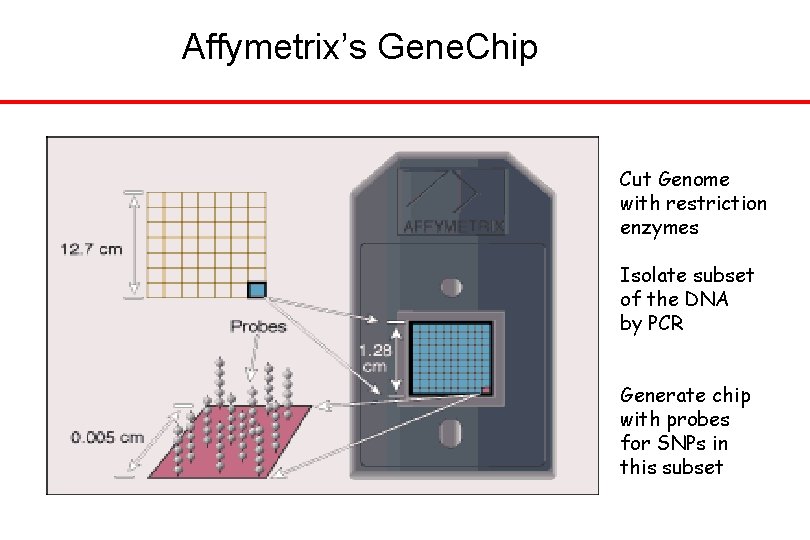
Affymetrix’s Gene. Chip Cut Genome with restriction enzymes Isolate subset of the DNA by PCR Generate chip with probes for SNPs in this subset
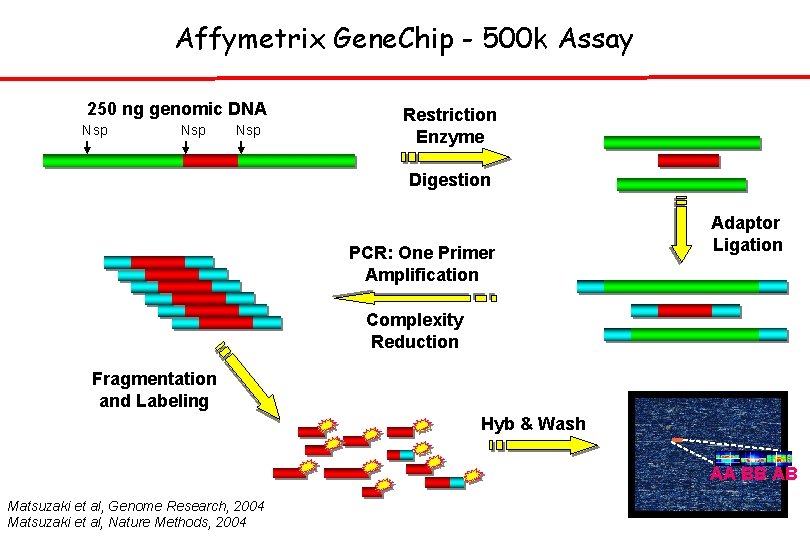
Affymetrix Gene. Chip - 500 k Assay 250 ng genomic DNA Nsp Nsp Restriction Enzyme Digestion PCR: One Primer Amplification Adaptor Ligation Complexity Reduction Fragmentation and Labeling Hyb & Wash AA BB AB Matsuzaki et al, Genome Research, 2004 Matsuzaki et al, Nature Methods, 2004
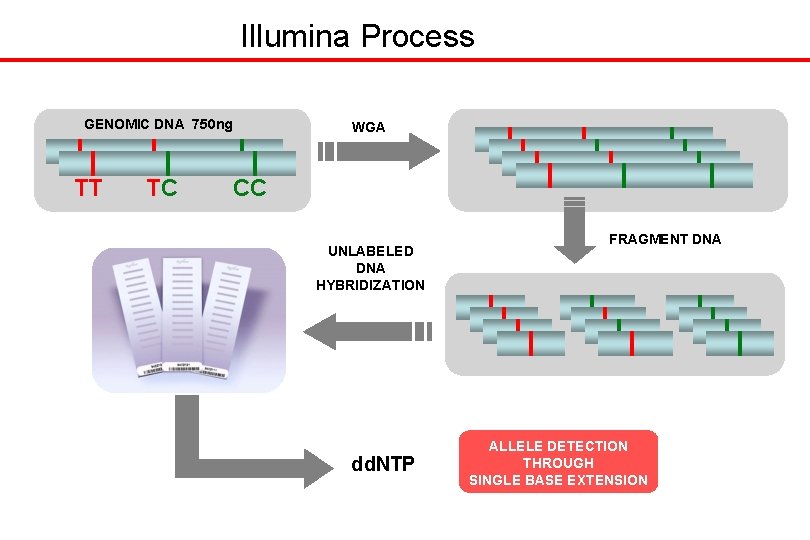
Illumina Process GENOMIC DNA 750 ng TT TC WGA CC UNLABELED DNA HYBRIDIZATION dd. NTP FRAGMENT DNA ALLELE DETECTION THROUGH SINGLE BASE EXTENSION
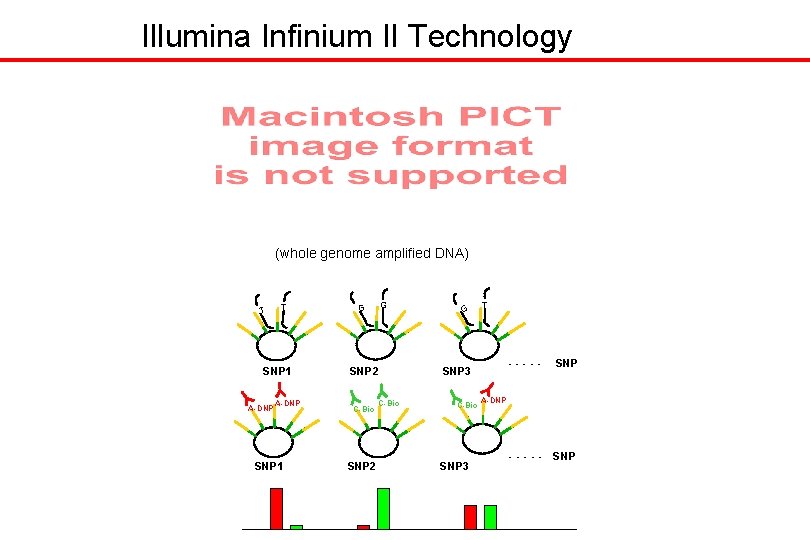
Illumina Infinium II Technology (whole genome amplified DNA) T T SNP 1 A-DNP SNP 1 G G SNP 2 C-Bio G T ----- SNP 3 C-Bio SNP 3 SNP A-DNP - - - SNP

Illumina Bead Chip Infinium II - Two-color assay
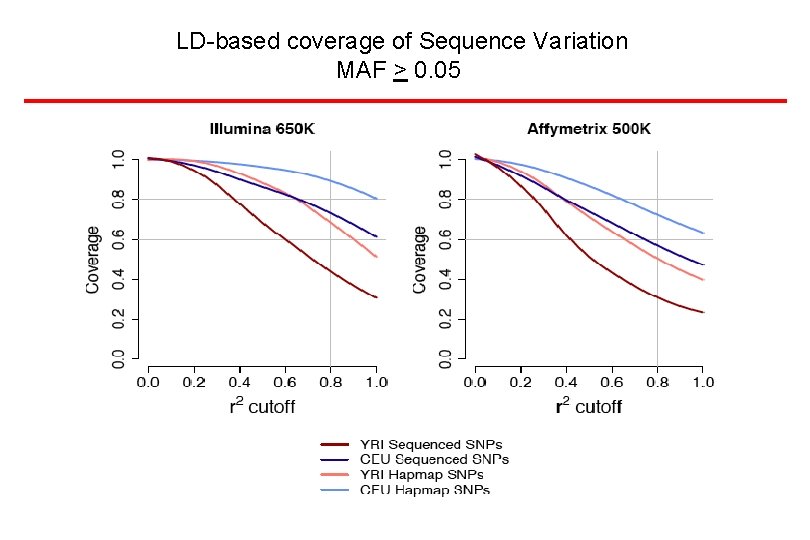
LD-based coverage of Sequence Variation MAF > 0. 05
Whole Genome Association Studies of Complex Traits Many Advantages –Detects common variation with small genetic contributions such as those in complex disease traits - where multiple genes are involved –Association defines a relatively small region (with hopefully one or few genes) –Does not require a priori knowledge of what genes or regions are involved Caveats –Requires thousands of samples to find a significant association –Extremely large datasets are generated (e. g. , 2000 samples X 500, 000 loci or more than 1 billion genotypes) – This is just the Start - Analysis and Replication Strategies Are Key The Hope The identified targets will lead to new biological and medical Insights and translate into new and improved treatments for common human diseases

Applying Genome Variation - Will it work? YES!! Hits: Macular Degeneration, Obesity, Cardiac Repolarization, Inflammatory Bowel Disease, Diabetes T 1 and T 2, Coronary Artery Disease, Rheumatoid Arthritis, Breast Cancer, Colon Cancer, Asthma …… - There are misses as well; unclear why - phenotype, coverage, environmental contexts? Example of a miss - Hypertension -There are lots more hits in these data sets - sample size, low proxy coverage with other SNPs …. . - Analysis of associations between phenotype(s, ) and even individual sites is daunting, and this will just be the first stage, and this does even consider multi-site interactions.

Genome-wide Tour de force Nature 447: 661 -678 Read all the supplemental materials too!
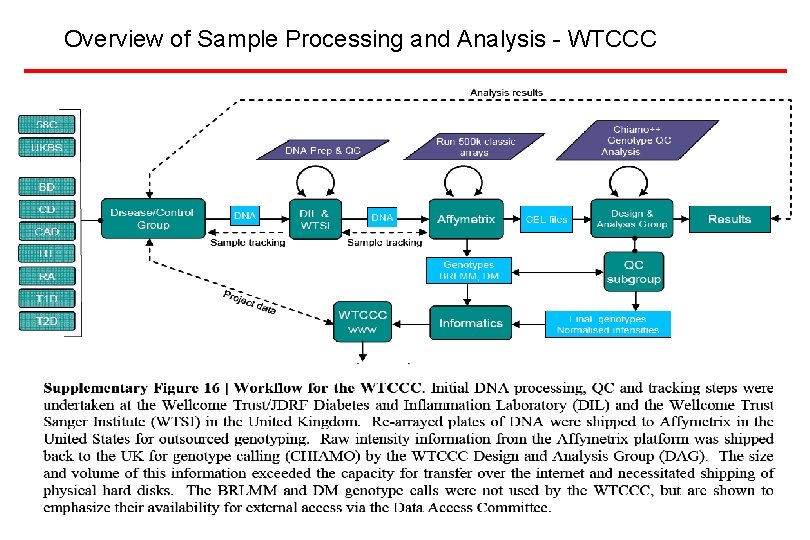
Overview of Sample Processing and Analysis - WTCCC

Data Quality Control • • Estimating Error Rates Hardy Weinberg Equilibrium Frequency Analysis Missing Data
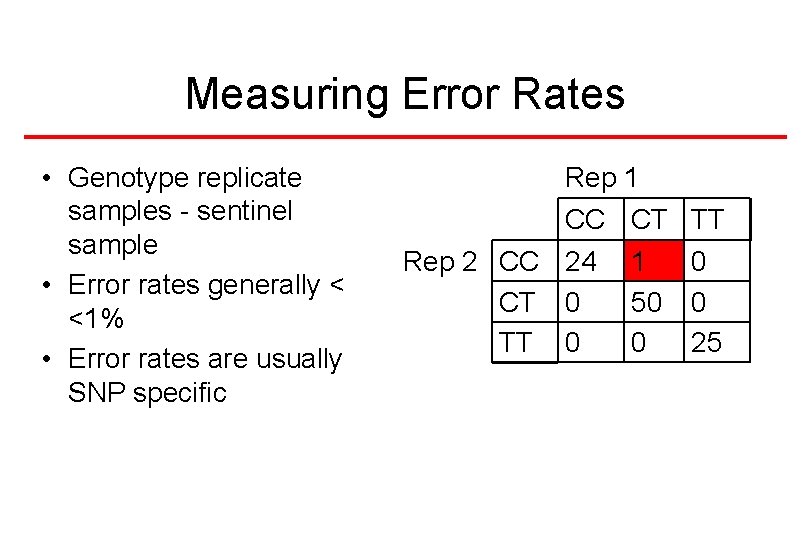
Measuring Error Rates • Genotype replicate samples - sentinel sample • Error rates generally < <1% • Error rates are usually SNP specific Rep 1 CC CT Rep 2 CC 24 1 CT 0 50 TT 0 0 25
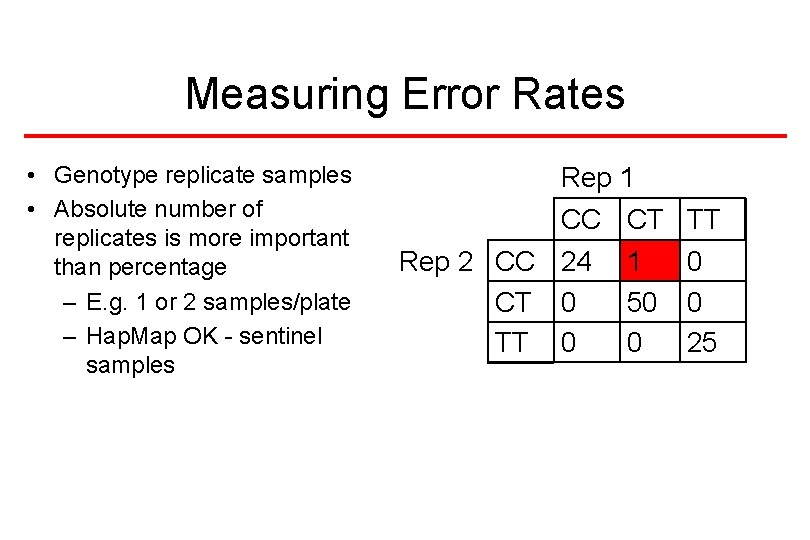
Measuring Error Rates • Genotype replicate samples • Absolute number of replicates is more important than percentage – E. g. 1 or 2 samples/plate – Hap. Map OK - sentinel samples Rep 1 CC CT Rep 2 CC 24 1 CT 0 50 TT 0 0 25
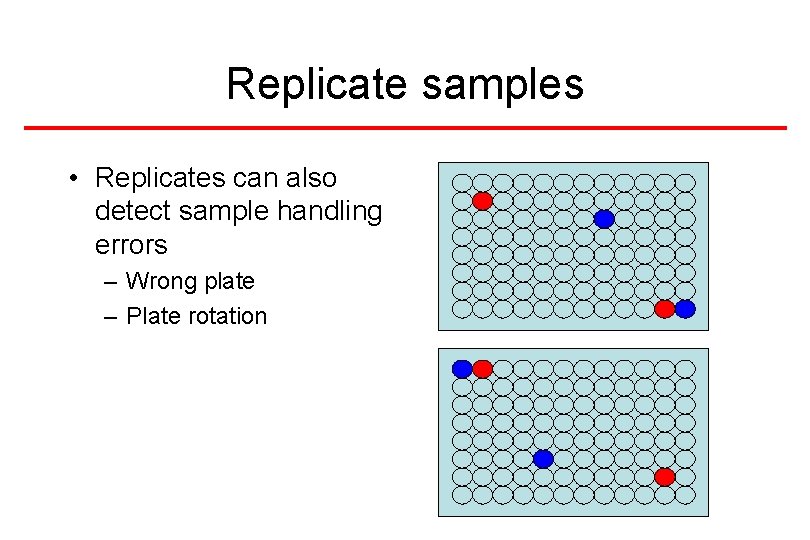
Replicate samples • Replicates can also detect sample handling errors – Wrong plate – Plate rotation
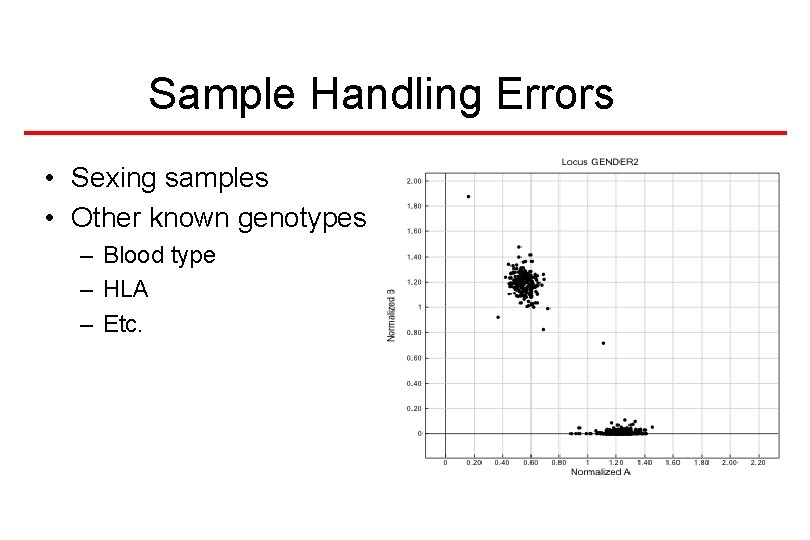
Sample Handling Errors • Sexing samples • Other known genotypes – Blood type – HLA – Etc.
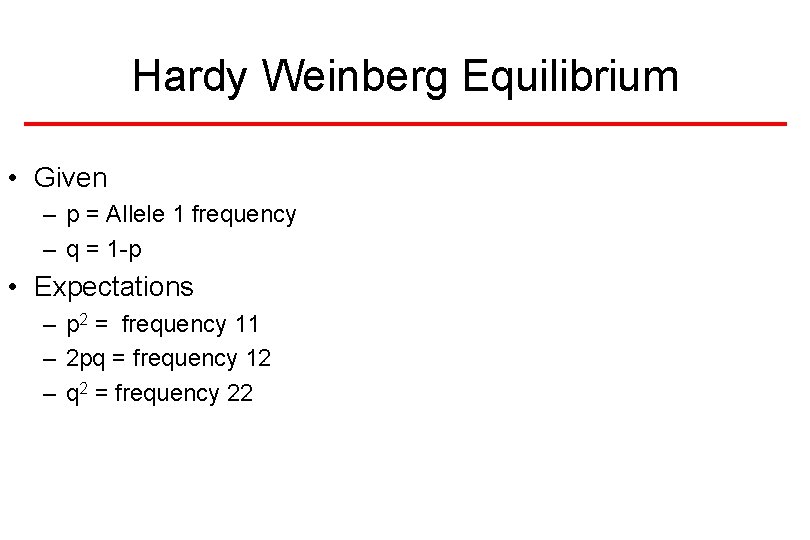
Hardy Weinberg Equilibrium • Given – p = Allele 1 frequency – q = 1 -p • Expectations – p 2 = frequency 11 – 2 pq = frequency 12 – q 2 = frequency 22
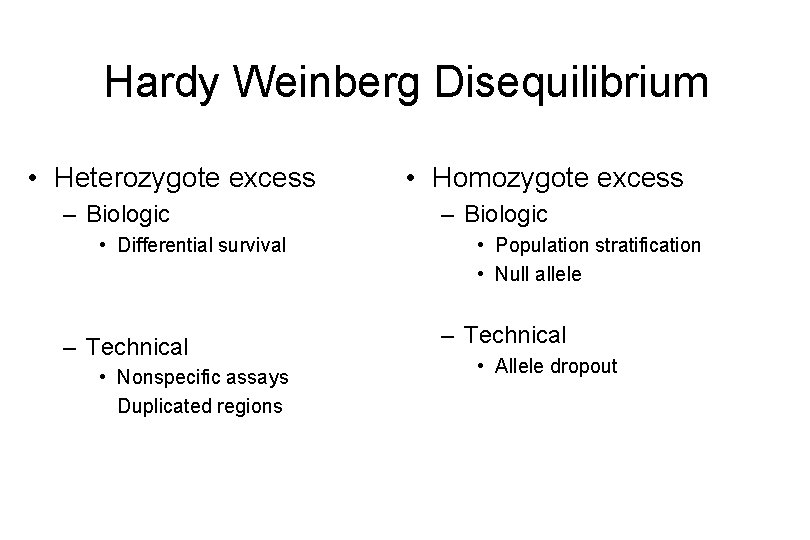
Hardy Weinberg Disequilibrium • Heterozygote excess – Biologic • Differential survival – Technical • Nonspecific assays Duplicated regions • Homozygote excess – Biologic • Population stratification • Null allele – Technical • Allele dropout
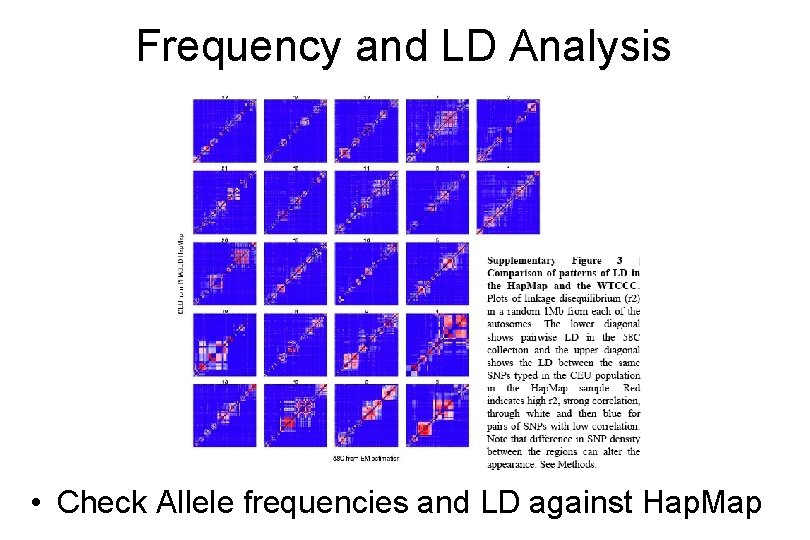
Frequency and LD Analysis European African • Check Allele frequencies and LD against Hap. Map
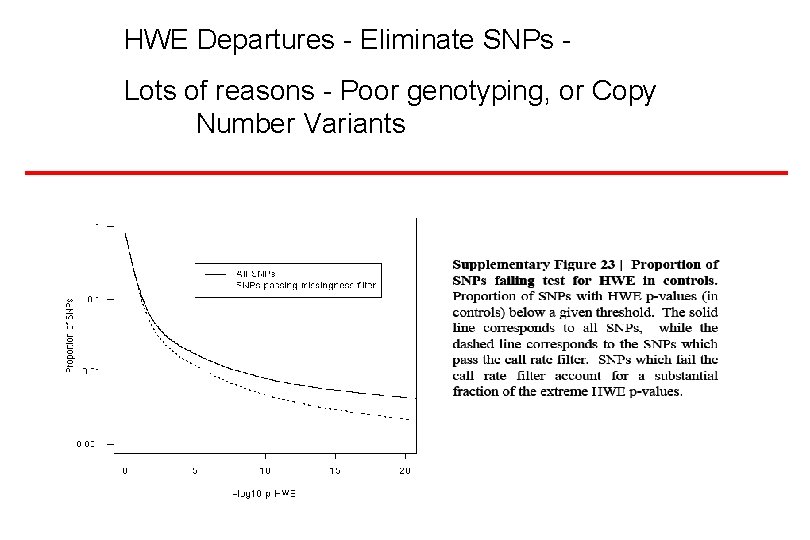
HWE Departures - Eliminate SNPs Lots of reasons - Poor genotyping, or Copy Number Variants
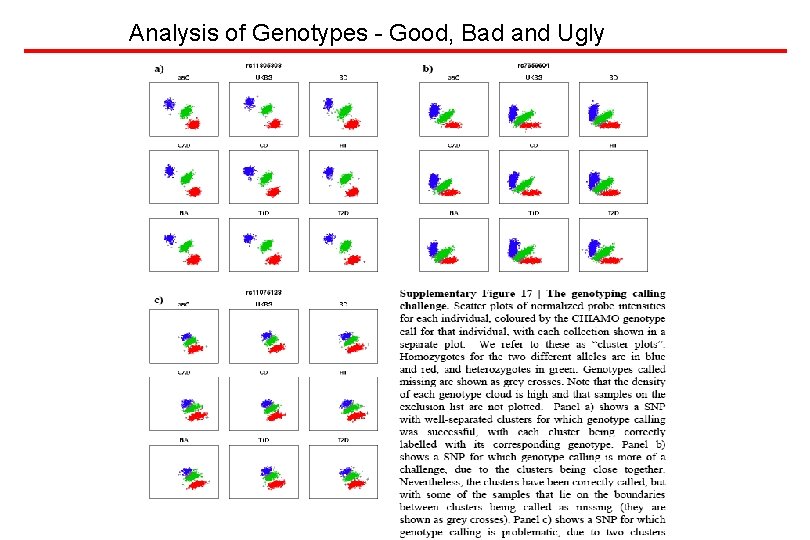
Analysis of Genotypes - Good, Bad and Ugly
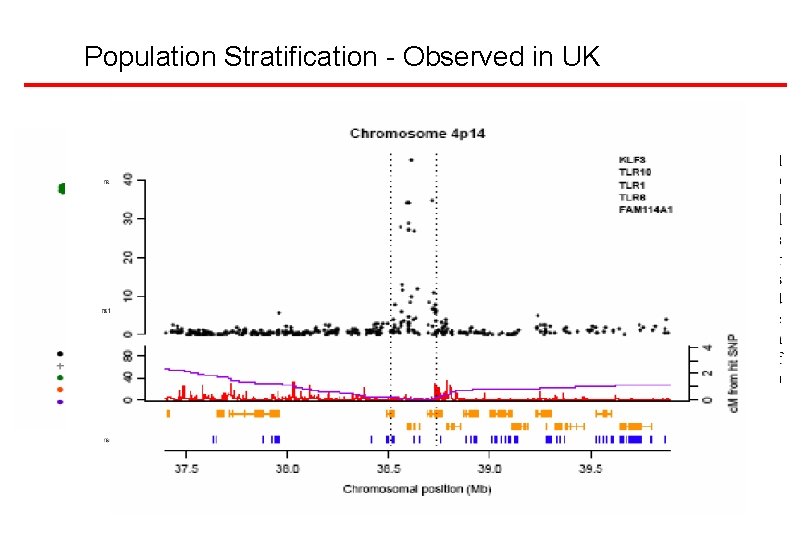
Population Stratification - Observed in UK
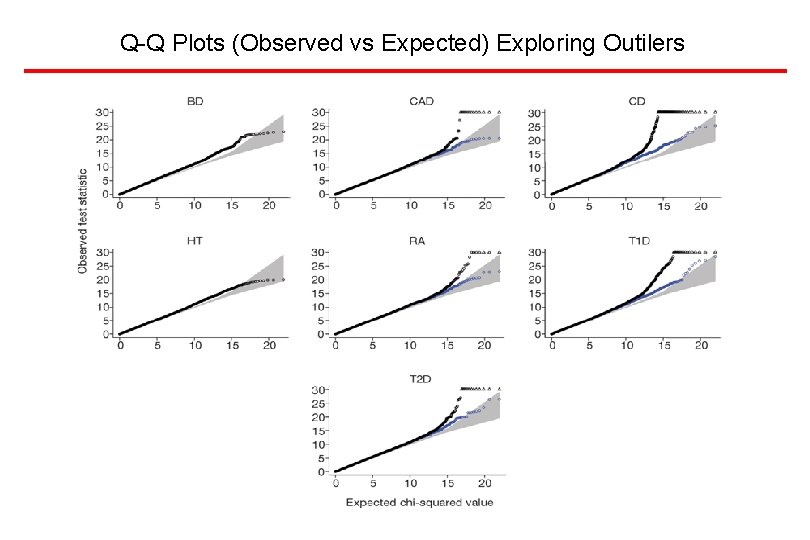
Q-Q Plots (Observed vs Expected) Exploring Outilers
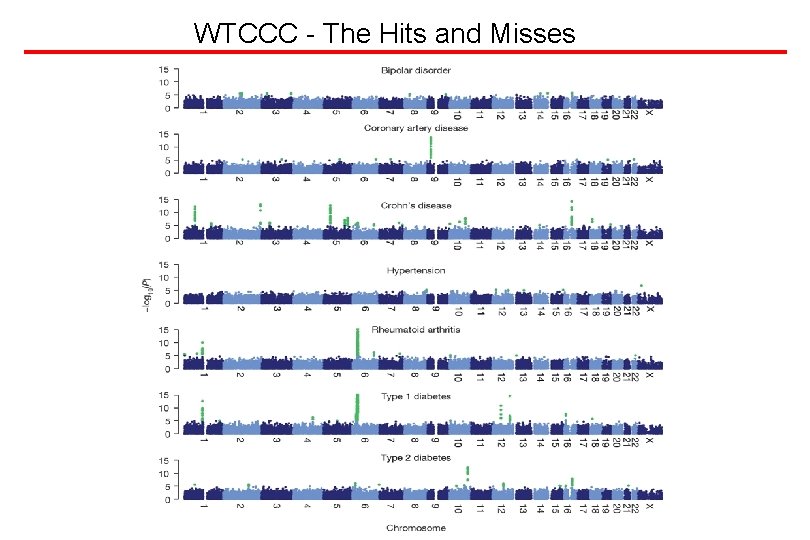
WTCCC - The Hits and Misses
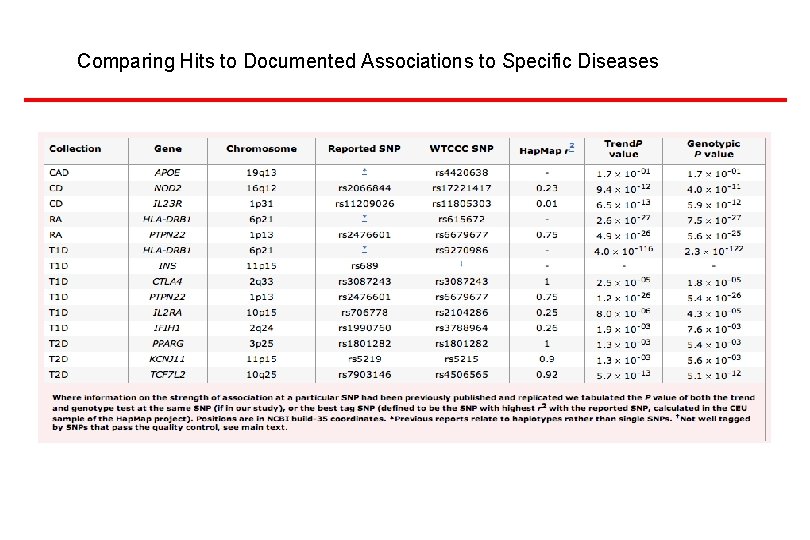
Comparing Hits to Documented Associations to Specific Diseases
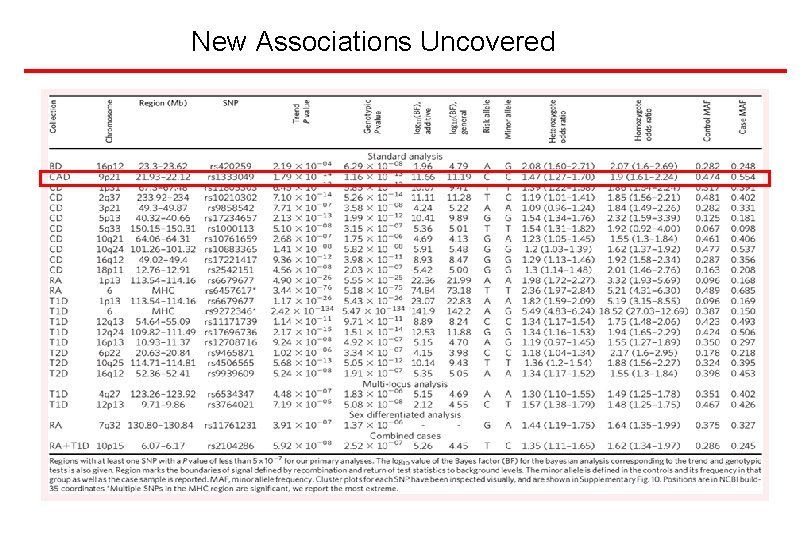
New Associations Uncovered
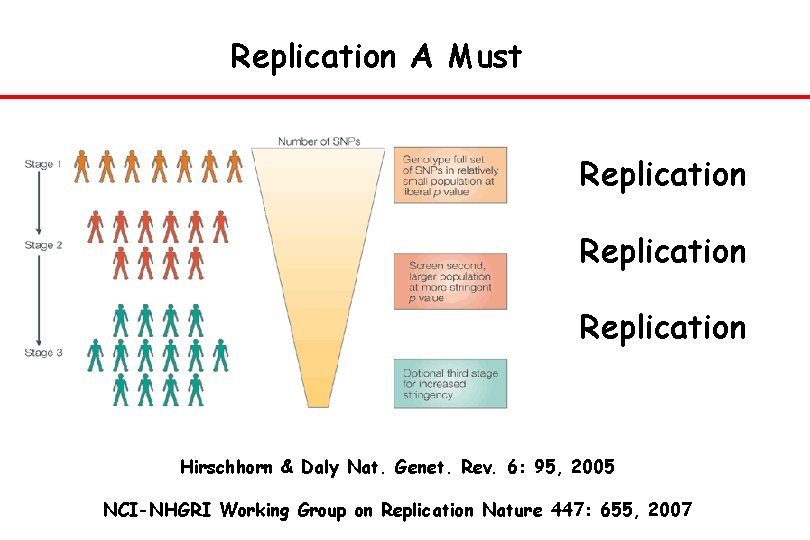
Replication A Must Replication Hirschhorn & Daly Nat. Genet. Rev. 6: 95, 2005 NCI-NHGRI Working Group on Replication Nature 447: 655, 2007
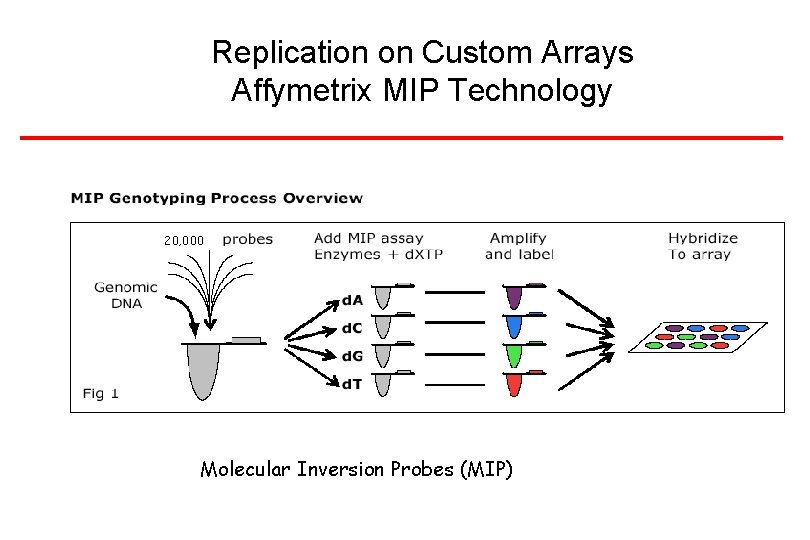
Replication on Custom Arrays Affymetrix MIP Technology 20, 000 Molecular Inversion Probes (MIP)
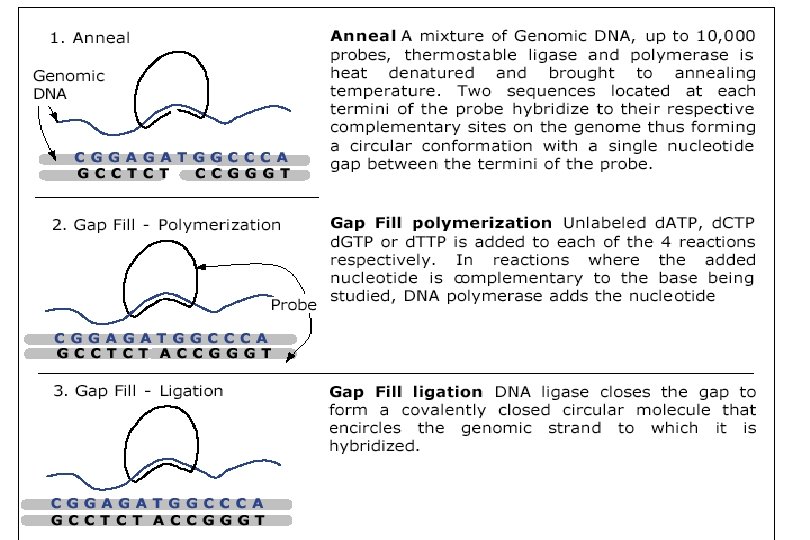
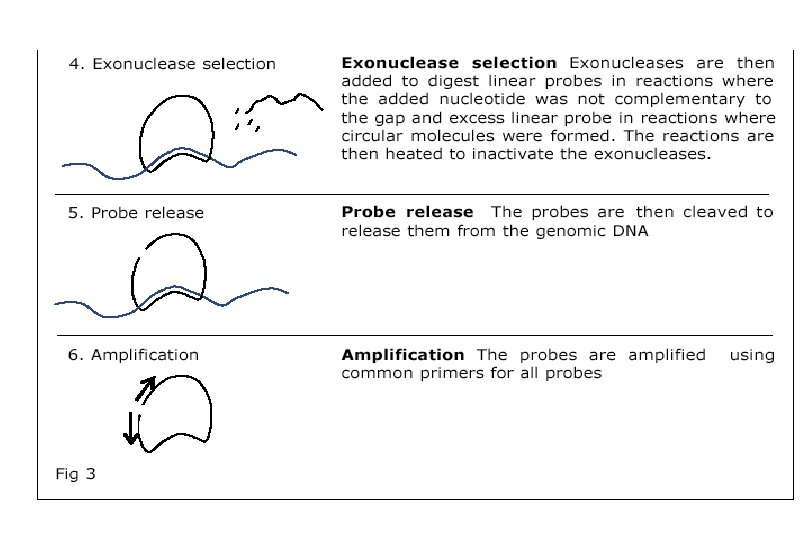
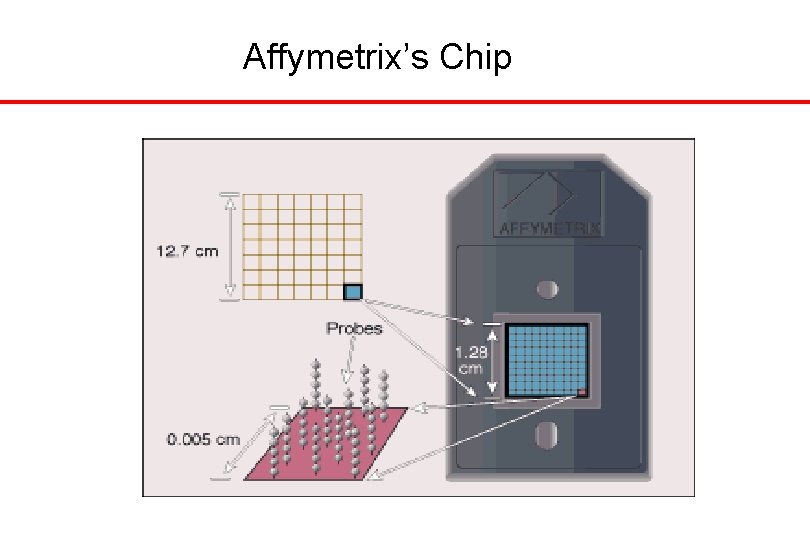
Affymetrix’s Chip

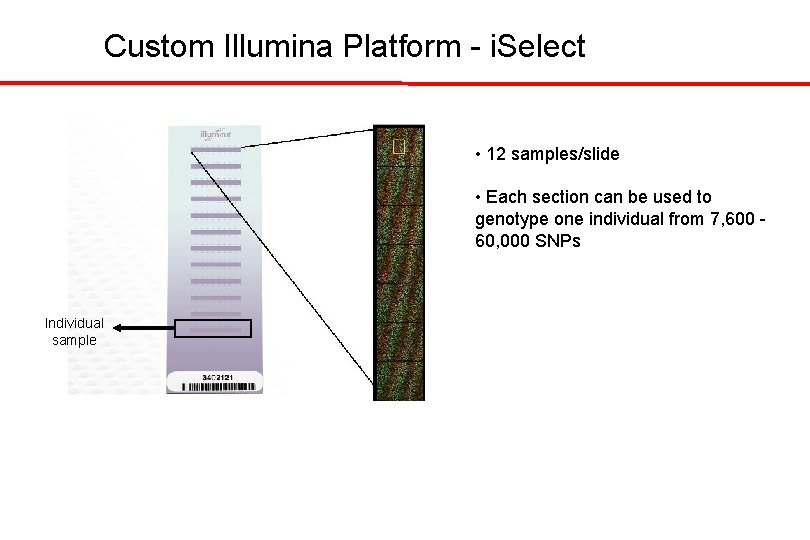
Custom Illumina Platform - i. Select • 12 samples/slide • Each section can be used to genotype one individual from 7, 600 60, 000 SNPs Individual sample
GWAS - CAD First report WTCCC Second
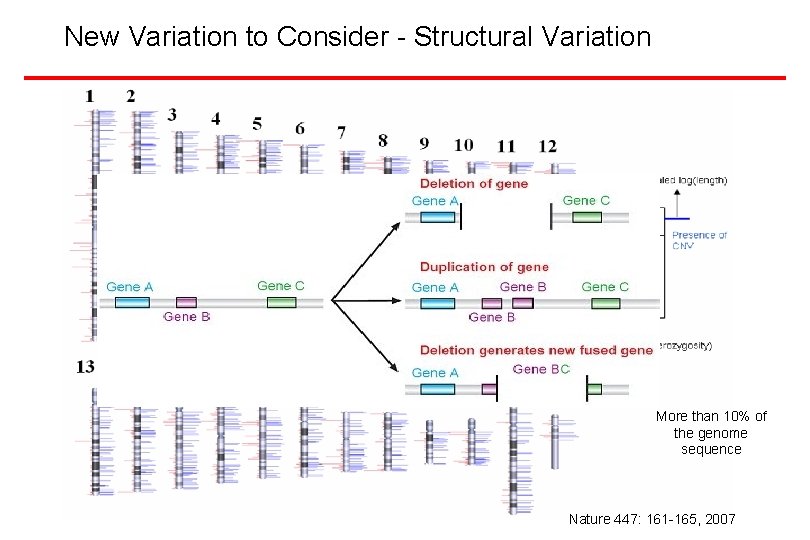
New Variation to Consider - Structural Variation Types of Structural Variants Insertions/Deletions Inversions Duplications Translocations Size: Large-scale (>100 kb) intermediate-scale (500 bp– 100 kb) Fine-scale (1– 500 bp) More than 10% of the genome sequence Nature 447: 161 -165, 2007
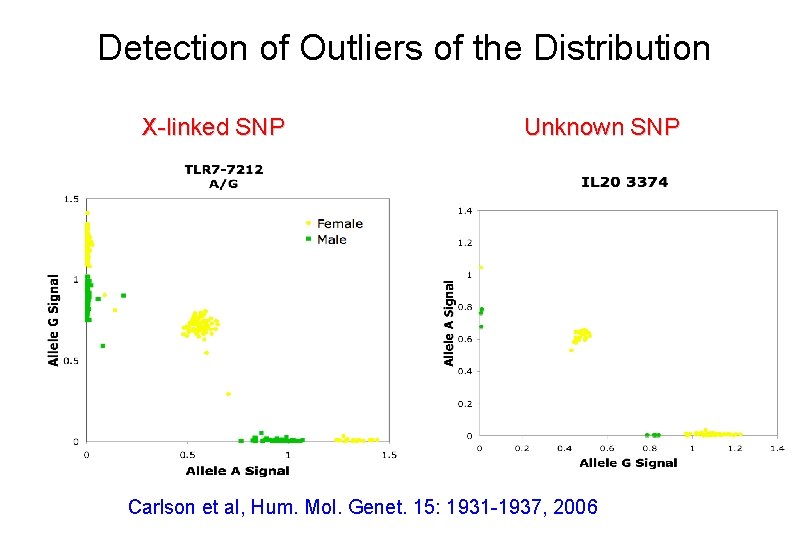
Detection of Outliers of the Distribution X-linked SNP Unknown SNP Carlson et al, Hum. Mol. Genet. 15: 1931 -1937, 2006

Structural Variation - Large Insertion-Deletion Events Structural Variants Identified in the Hap. Map • Conrad, et al. (Nature Genetics 38: 75 -81, 2006) • Hinds, et al. (Nature Genetics 38: 82 -85, 2006) • Mc. Carroll, et al. (Nature Genetics 38: 86 -92, 2006) Nearly 4, 000 now known
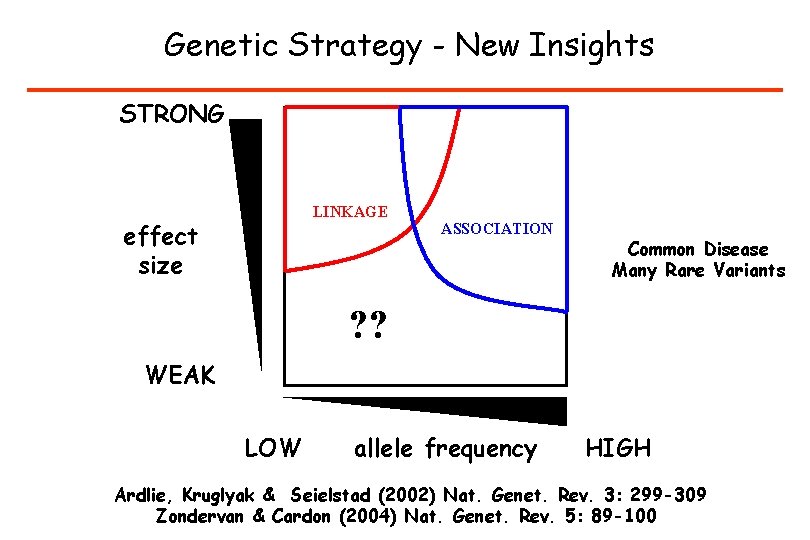
Genetic Strategy - New Insights STRONG LINKAGE effect size ASSOCIATION Common Disease Many Rare Variants ? ? WEAK LOW allele frequency HIGH Ardlie, Kruglyak & Seielstad (2002) Nat. Genet. Rev. 3: 299 -309 Zondervan & Cardon (2004) Nat. Genet. Rev. 5: 89 -100
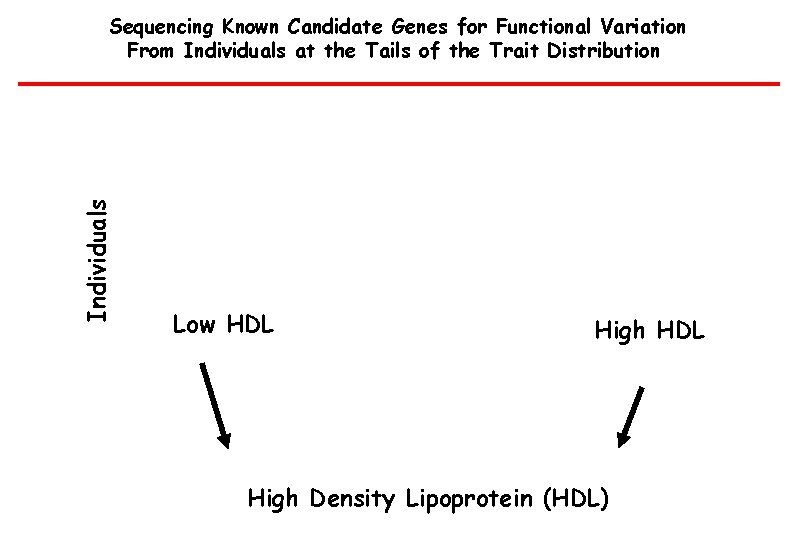
Individuals Sequencing Known Candidate Genes for Functional Variation From Individuals at the Tails of the Trait Distribution Low HDL High Density Lipoprotein (HDL)
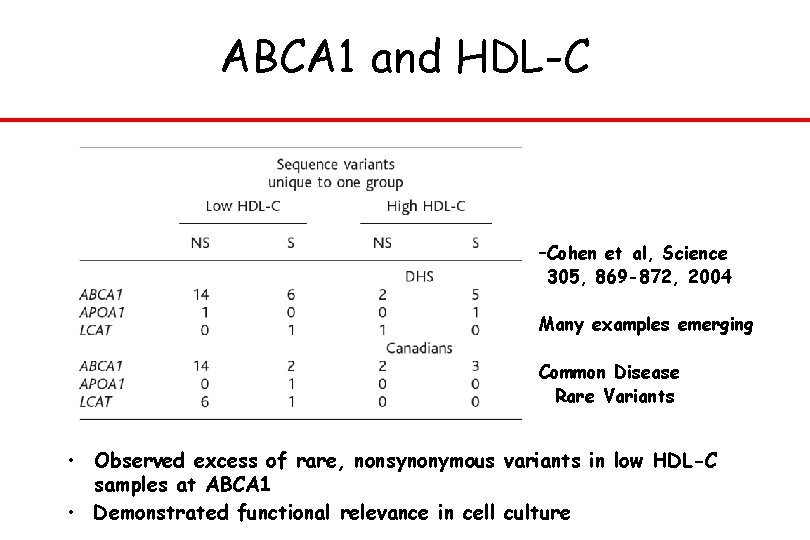
ABCA 1 and HDL-C –Cohen et al, Science 305, 869 -872, 2004 Many examples emerging Common Disease Rare Variants • Observed excess of rare, nonsynonymous variants in low HDL-C samples at ABCA 1 • Demonstrated functional relevance in cell culture
Personalized Human Genome Sequencing Solexa - an example
Genotyping Summary • Hap. Map - the spectrum of common variation • New Genotyping Platforms - Not perfect but successful • Uncovering Many Associations - Many regions underlying common disease associations uncovered - WTCC as a paradigm • Stratification is being uncovered on many levels - within and between populations and these generate artificial associations • Replication is a must to explore genome-wide associations • New technologies for replication on large-scale • New variants and sequencing technologies emerging
- Slides: 71