Overview of Microarray Gene Expression 2 n Gene

- Slides: 31

Overview of Microarray

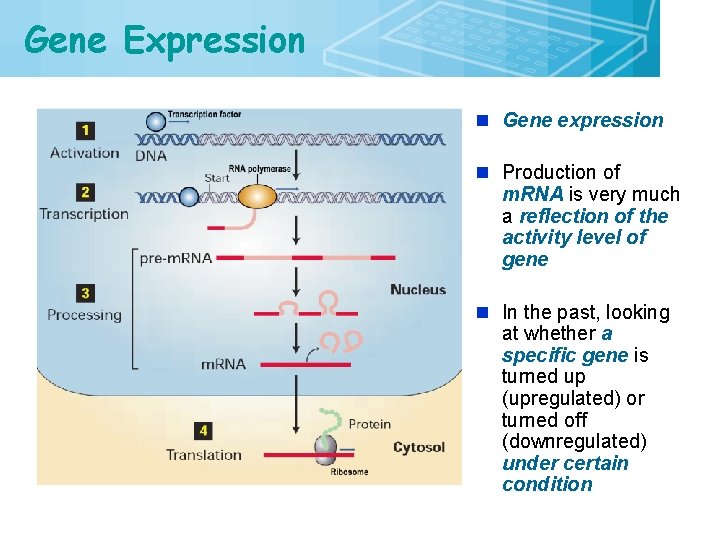
Gene Expression 2 n Gene expression n Production of m. RNA is very much a reflection of the activity level of gene n In the past, looking at whether a specific gene is turned up (upregulated) or turned off (downregulated) under certain condition

Microarray Data n Microarray: a technological advancement study the genes of an organism’s at once n Microarrays are a massively-parallel Northern Blot n High throughput method allow for the global study of changes in gene expression → complete cellular snapshot 3

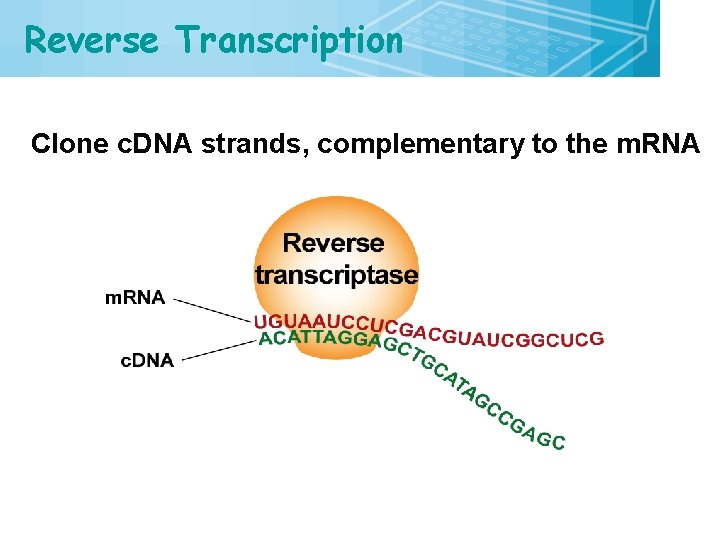
Reverse Transcription 4 Clone c. DNA strands, complementary to the m. RNA

Microarray Experiments m. RNA levels compared in many different contexts Different tissues, same organism (brain vs. liver) Same tissue, same organism (tumor vs. non-tumor) Same tissue, different organisms (wild-type or mutant) Time course experiments (development) 5

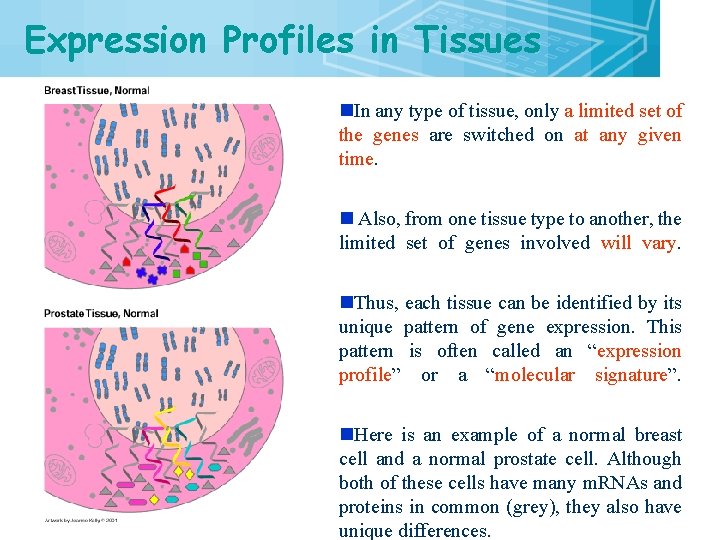
Expression Profiles in Tissues n. In any type of tissue, only a limited set of the genes are switched on at any given time. n Also, from one tissue type to another, the limited set of genes involved will vary. n. Thus, each tissue can be identified by its unique pattern of gene expression. This pattern is often called an “expression profile” or a “molecular signature”. n. Here is an example of a normal breast cell and a normal prostate cell. Although both of these cells have many m. RNAs and proteins in common (grey), they also have unique differences. 6

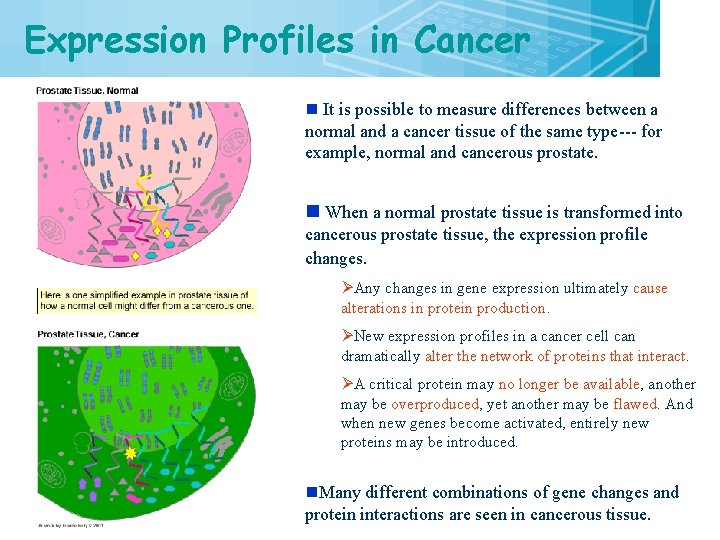
Expression Profiles in Cancer 7 n It is possible to measure differences between a normal and a cancer tissue of the same type--- for example, normal and cancerous prostate. n When a normal prostate tissue is transformed into cancerous prostate tissue, the expression profile changes. ØAny changes in gene expression ultimately cause alterations in protein production. ØNew expression profiles in a cancer cell can dramatically alter the network of proteins that interact. ØA critical protein may no longer be available, another may be overproduced, yet another may be flawed. And when new genes become activated, entirely new proteins may be introduced. n. Many different combinations of gene changes and protein interactions are seen in cancerous tissue.

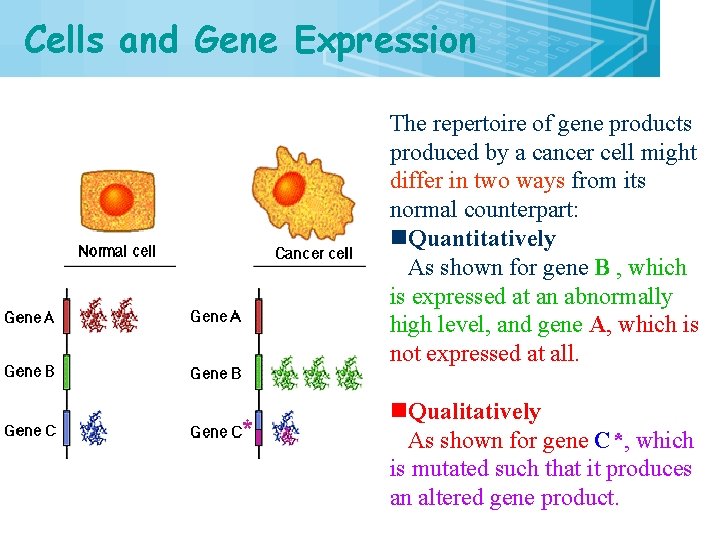
Cells and Gene Expression 8 The repertoire of gene products produced by a cancer cell might differ in two ways from its normal counterpart: n. Quantitatively As shown for gene B , which is expressed at an abnormally high level, and gene A, which is not expressed at all. n. Qualitatively As shown for gene C*, which is mutated such that it produces an altered gene product.

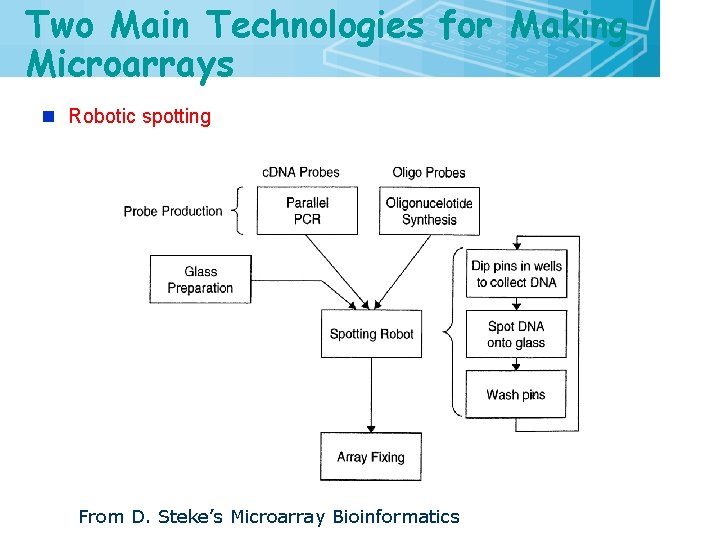
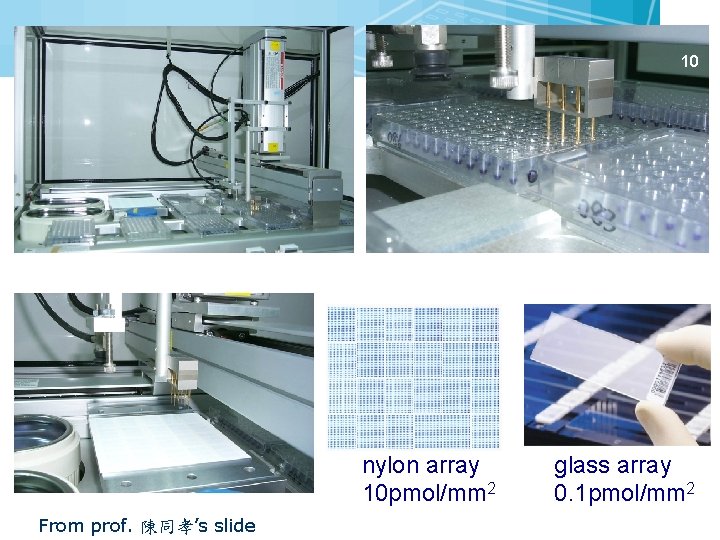
Two Main Technologies for Making Microarrays n Robotic spotting From D. Steke’s Microarray Bioinformatics 9

10 nylon array 10 pmol/mm 2 From prof. 陳同孝’s slide glass array 0. 1 pmol/mm 2

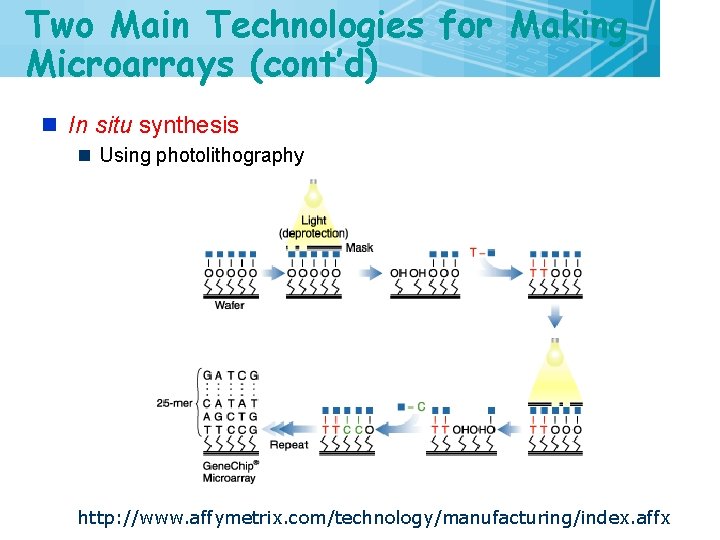
Two Main Technologies for Making Microarrays (cont’d) n In situ synthesis n Using photolithography http: //www. affymetrix. com/technology/manufacturing/index. affx 11

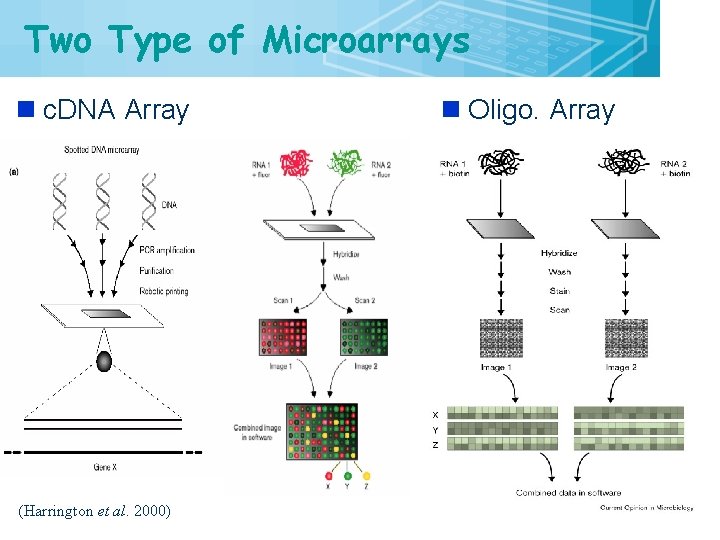
Two Type of Microarrays n c. DNA Array (Harrington et al. 2000) n Oligo. Array 12

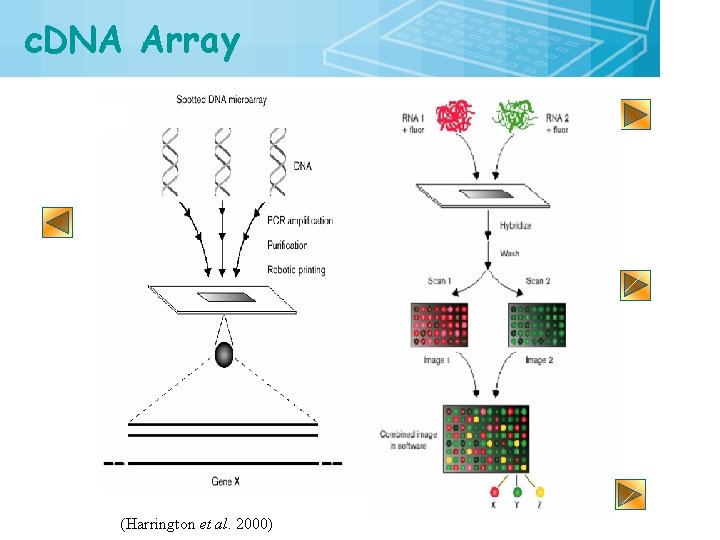
c. DNA Array (Harrington et al. 2000) 13

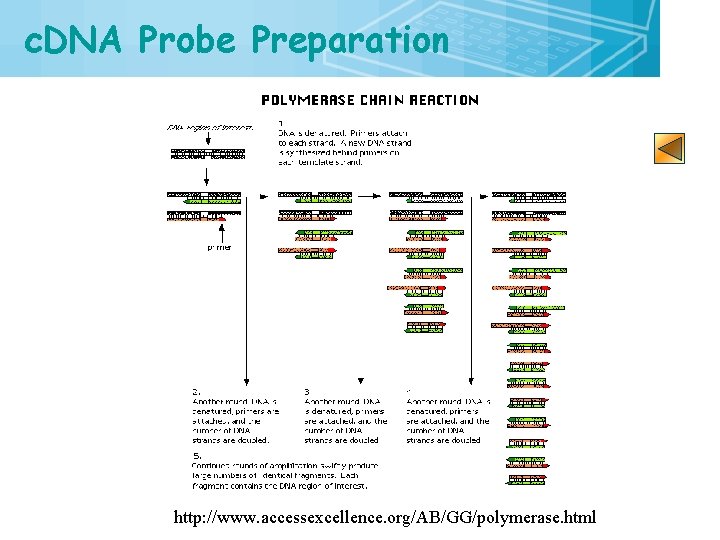
c. DNA Probe Preparation http: //www. accessexcellence. org/AB/GG/polymerase. html 14

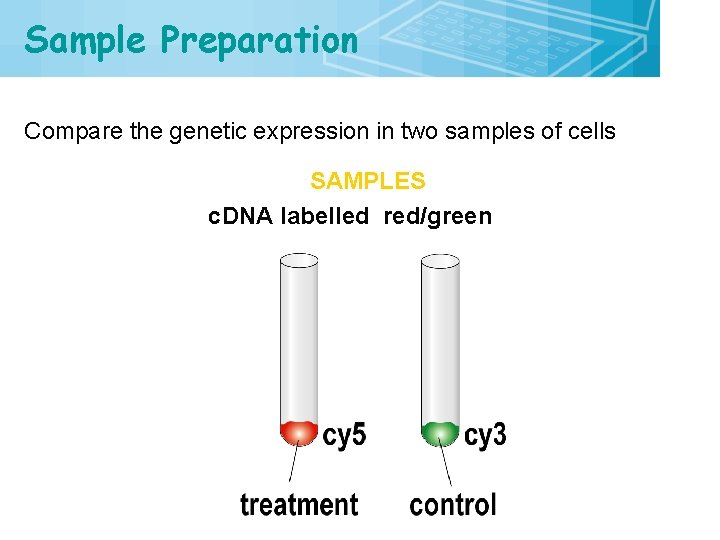
Sample Preparation Compare the genetic expression in two samples of cells SAMPLES c. DNA labelled red/green 15

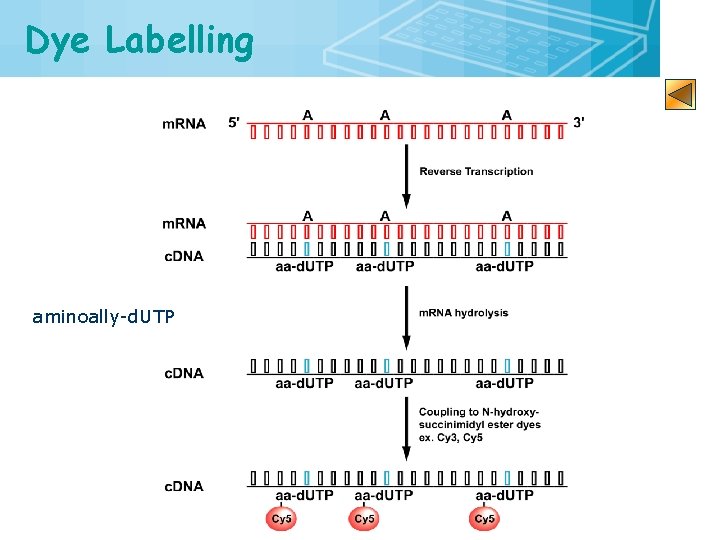
Dye Labelling aminoally-d. UTP 16

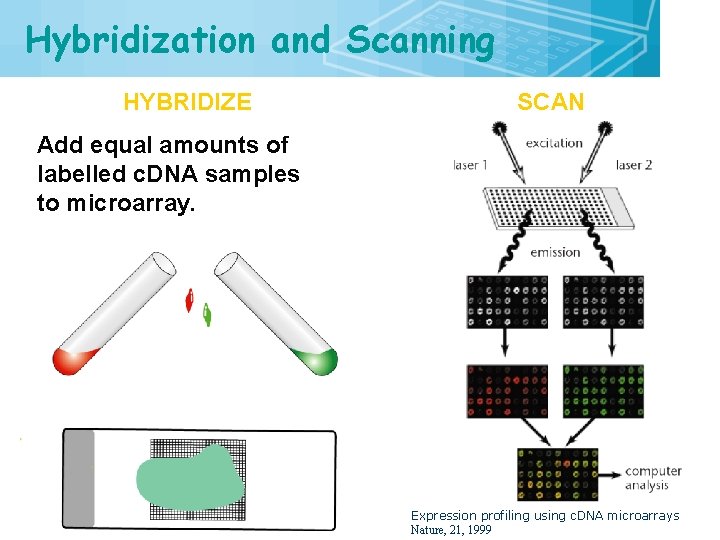
Hybridization and Scanning HYBRIDIZE 17 SCAN Add equal amounts of labelled c. DNA samples to microarray. Expression profiling using c. DNA microarrays Nature, 21, 1999

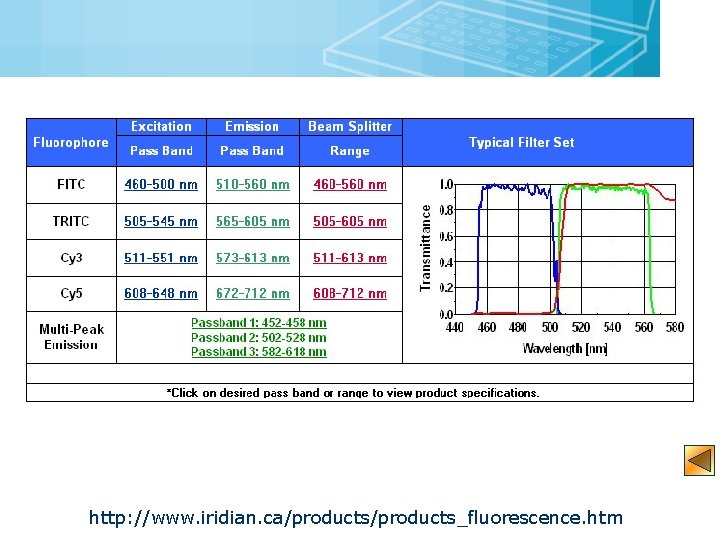
18 http: //www. iridian. ca/products_fluorescence. htm

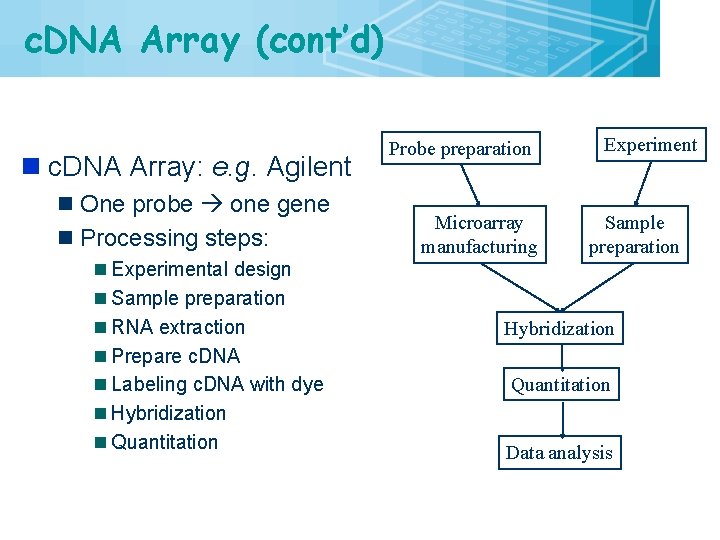
c. DNA Array (cont’d) n c. DNA Array: e. g. Agilent n One probe one gene n Processing steps: n Experimental design n Sample preparation n RNA extraction n Prepare c. DNA n Labeling c. DNA with dye n Hybridization n Quantitation 19 Probe preparation Microarray manufacturing Experiment Sample preparation Hybridization Quantitation Data analysis

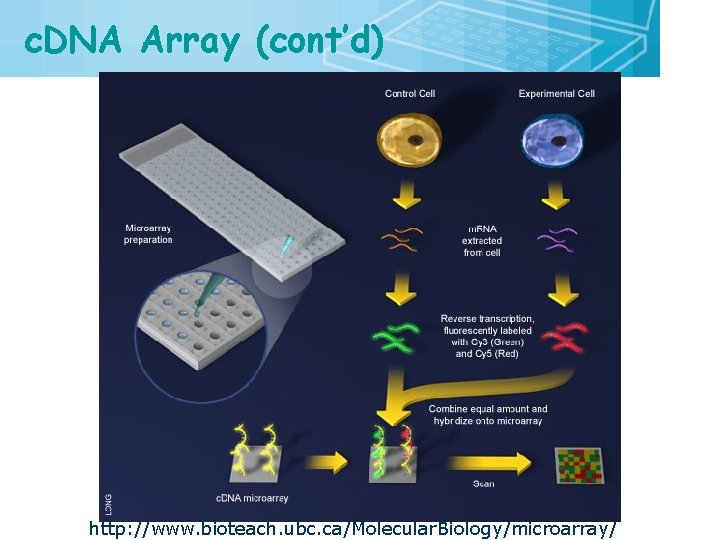
c. DNA Array (cont’d) http: //www. bioteach. ubc. ca/Molecular. Biology/microarray/ 20

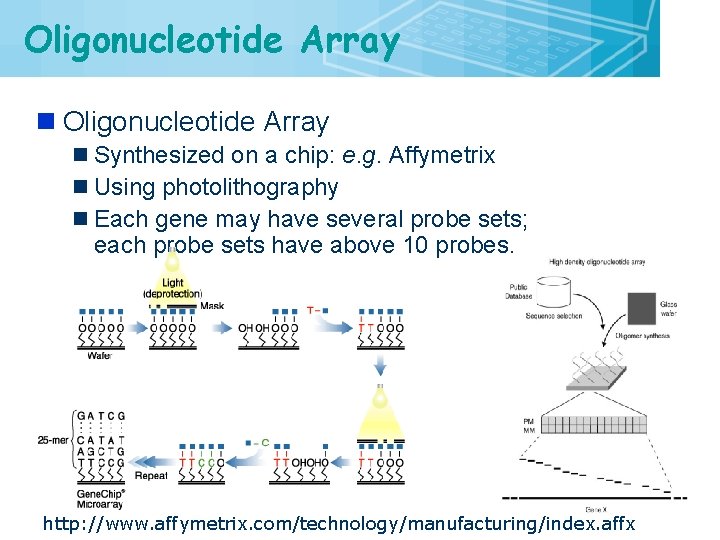
Oligonucleotide Array n Synthesized on a chip: e. g. Affymetrix n Using photolithography n Each gene may have several probe sets; each probe sets have above 10 probes. http: //www. affymetrix. com/technology/manufacturing/index. affx 21

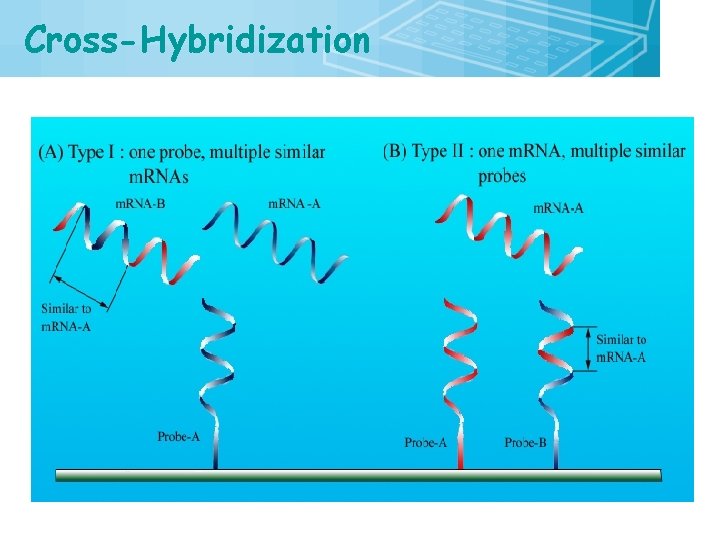
Cross-Hybridization 22

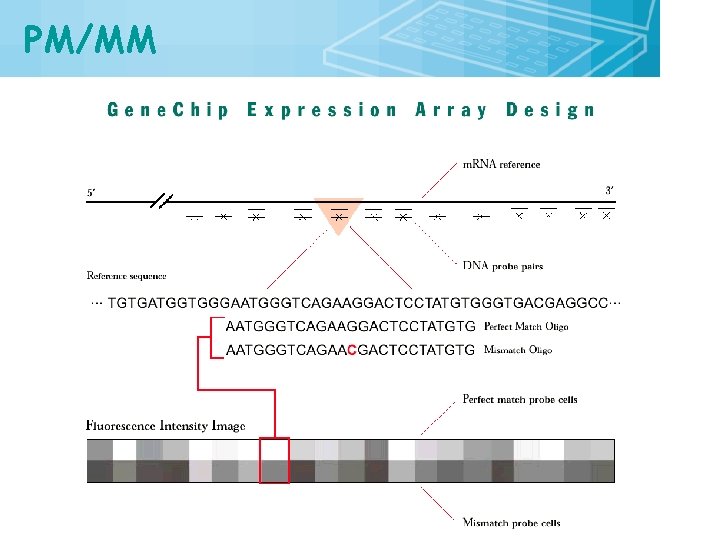
PM/MM 23

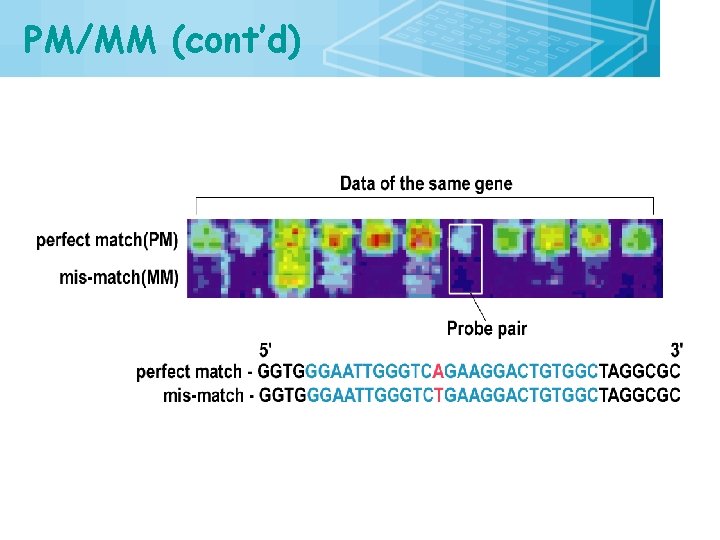
PM/MM (cont’d) 24

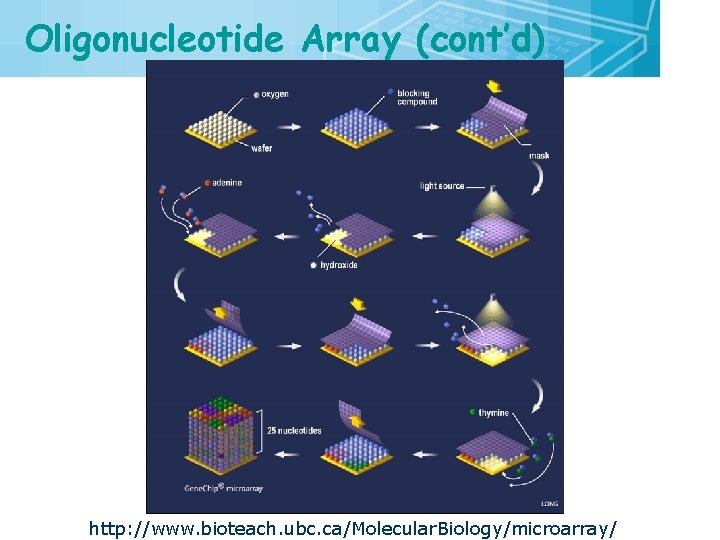
Oligonucleotide Array (cont’d) http: //www. bioteach. ubc. ca/Molecular. Biology/microarray/ 25

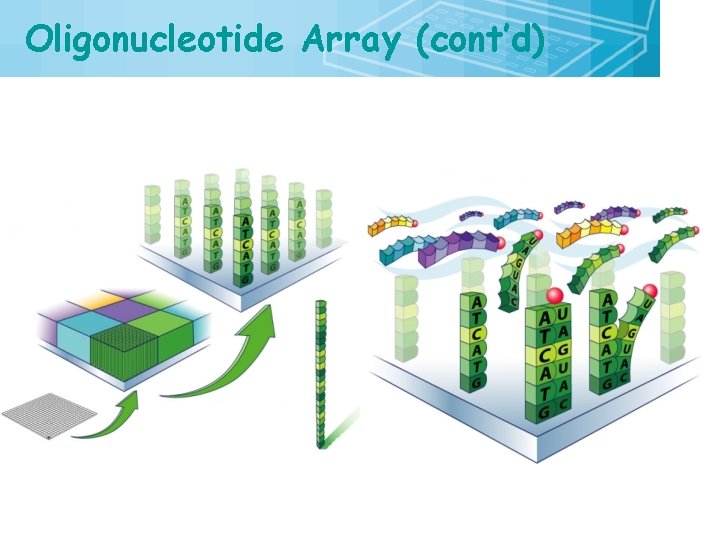
Oligonucleotide Array (cont’d) 26

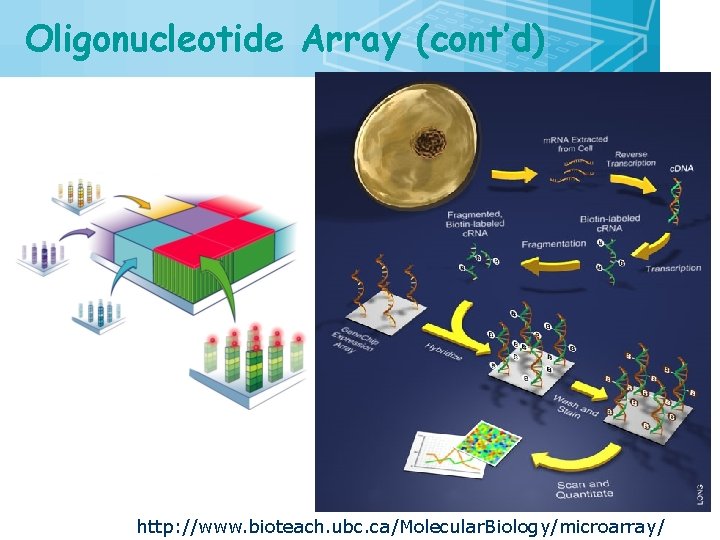
Oligonucleotide Array (cont’d) http: //www. bioteach. ubc. ca/Molecular. Biology/microarray/ 27

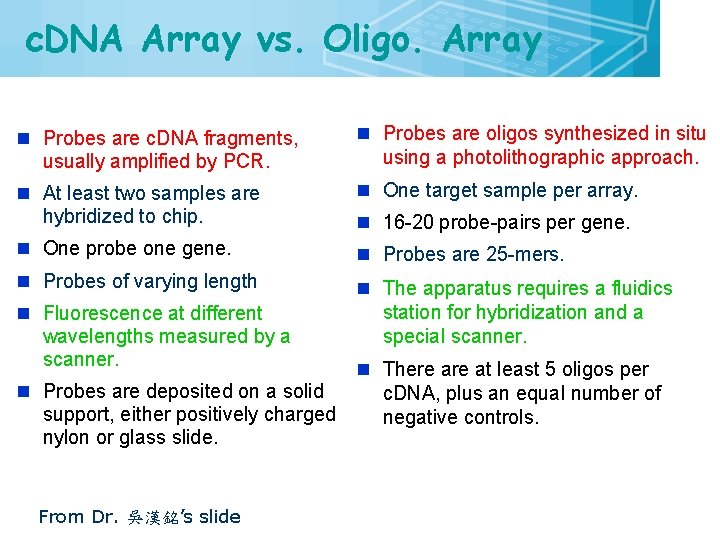
c. DNA Array vs. Oligo. Array 28 n Probes are c. DNA fragments, usually amplified by PCR. n Probes are oligos synthesized in situ using a photolithographic approach. n At least two samples are hybridized to chip. n One target sample per array. n One probe one gene. n Probes are 25 -mers. n Probes of varying length n The apparatus requires a fluidics station for hybridization and a special scanner. n Fluorescence at different wavelengths measured by a scanner. n Probes are deposited on a solid support, either positively charged nylon or glass slide. From Dr. 吳漢銘’s slide n 16 -20 probe-pairs per gene. n There at least 5 oligos per c. DNA, plus an equal number of negative controls.

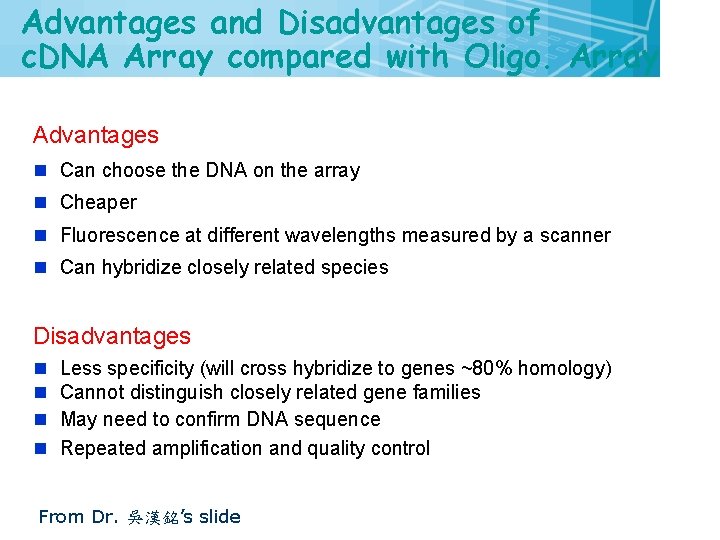
Advantages and Disadvantages of c. DNA Array compared with Oligo. Array Advantages n Can choose the DNA on the array n Cheaper n Fluorescence at different wavelengths measured by a scanner n Can hybridize closely related species Disadvantages n Less specificity (will cross hybridize to genes ~80% homology) n Cannot distinguish closely related gene families n May need to confirm DNA sequence n Repeated amplification and quality control From Dr. 吳漢銘’s slide 29

Advantages and Disadvantages of c. DNA Array compared with Oligo. Array 30 Advantages n High specificity (small probe length means gene family members can be differentiated) n Very robust protocols and results are very reproducible n Can use small amount of RNA n Widely used, so annotation of probe sets is of relatively high quality Disadvantages n Very expensive to design (~US$300, 000) n Expensive to perform experiments (~US$400 + $300 labeling/hybridization) n Limited to the species for which there are chips available sequence required n Single target hybridization, so comparison always involves two experiments, and dye swaps are impossible n Match/mismatch technology has major limitations: mismatch signal often higher than match, and dose response curve is different for each pair From Dr. 吳漢銘’s slide

Microarray Experimental Flowchart 31 Microarray Life Cycle R=Rf-Rb G=Gf-Gb M=log 2 R/G A=1/2 log 2 RG